
Medical Bioinformatics and Computational Modelling
PhD students at the Bioinformatics Laboratory
In Progress
- Lashgari, D. Kinetic maturation in the Germinal Center . University of Amsterdam, Amsterdam. Supported by AMC. Van Kampen, A.H.C. (promotor), Van Gils, M. (co-promotor), Hoefsloot, H. C. (co-promotor).
- Mahamune, U. Single Cell RNAseq and computational modelling . University of Amsterdam, Amsterdam. ARCAID . Marie Curie COFUND, Horizon 2020. Van Kampen, A.H.C. (promotor), Moerland, P.D. (co-promotor), E.G.M. van Baarsen (co-promotor).
- Valiente, R. G. Development of multiscale mathematical models of the germinal center (GC) to study its role in B-cell lymphoma (BCL) and/or rheumatoid arthritis (RA). (PhD thesis). University of Amsterdam, Amsterdam. COSMIC . Marie Curie ITN, Horizon 2020. Van Kampen, A.H.C. (promotor), De Vries, N. (promotor), Hoefsloot, H. C. (co-promotor), Guikema, J. E. (co-promotor).
- Stobbe, M. (2012). 18 October 2012. The road to knowledge: from biology to databases and back again. University of Amsterdam, Amsterdam. NBIC BioRange. Van Kampen, A.H.C. (promotor), Moerland, P. D. (co-promotor). [ UvA-DARE ]
- Shahand, S. (2015). 29 October 2015. Science gateways for biomedical big data analysis. University of Amsterdam, Amsterdam. COMMIT. Van Kampen, A. (promotor), Olabarriaga, S. (co-promotor). [ UvA-DARE ]
- Reshetova, P. (2017). 2 March 2017. Use of Prior Knowledge in Biological Systems Modelling. University of Amsterdam, Amsterdam. NBIC Biorange. Van Kampen, A.H.C (promotor), Smilde, A. (promotor), Westerhuis, J. (co-promotor). [ UvA-DARE ]
- Tejero Merino, E. (2022). 7 November 2022 Multiscale modelling of plasma cell differentiation in the Germinal Center. University of Amsterdam, Amsterdam. Supported by AMC. Van Kampen, A.H.C. (promotor), Guikema, J.E.J. (co-promotor), Hoefsloot, H. C. (co-promotor). [ PhD thesis] [ UvA-DARE ]
- Nandal, U. (2023). Computational approaches for biological data integration. University of Amsterdam, Amsterdam. NBIC BioRange. Van Kampen, A.H.C. (promotor), Moerland, P.D. (co-promotor). [ UvA-DARE ]
- Balashova, D. Repertoire sequencing . University of Amsterdam, Amsterdam. ARCAID . Marie Curie COFUND, Horizon 2020. Van Kampen, A.H.C. (promotor), De Vries N. (promotor), Greiff V. (co-promotor). – Terminated
Co-supervised PhD students from other research groups
In Progress
- Balzaretti, G. Repertoire Sequencing . University of Amsterdam, Amsterdam. De Vries, N. (promotor), Van Kampen, A.H.C. (promotor).
- Lermo Jimenez, M. Epigenetics and breast cancer drug resistance . University of Amsterdam, Amsterdam. Verschure P. J. (promotor), Moerland, P.D. (co-promotor).
- Olivieri, A. Repertoire Sequencing. University of Amsterdam, Amsterdam. ARCAID , Marie Curie COFUND, Horizon 2020. De Vries, N. (promotor), Van Kampen, A.H.C. (promotor).
- Stratigopoulou, M. Germinal Center and B-cell Lymphoma . University of Amsterdam, Amsterdam. COSMIC. Marie Curie ITN, Horizon 2020. Van Kampen, A.H.C. (promotor), Van Noesel, C. J. (promotor), De Vries, N. (co- promotor), Guikema, J. E. (co-promotor).
- Sontrop, H. (2015). 15 January 2015. A critical perspective on microarray breast cancer gene expression profiling. TU Delft, Delft. NBIC BioRange. Reinders, M. (promotor), Moerland, P. D. (co-promotor). [ Link ]
- Beckman, W. (2021). 17 August 2021. The Role of Epigenetics in Transcriptional Stochasticity and the Implications for Breast Cancer Drug Resistance . University of Amsterdam, Amsterdam. EpiPredict. Marie Curie ITN, Horizon 2016. Verschure P.J. (promotor), Van Kampen, A.H.C. (promotor). [ UvA-DARE ]
- Barros, R. S. (2022). 1 November 2022 High performance computing for clinical medical imaging . University of Amsterdam, Amsterdam. Henk Marquering (promotor), Van Kampen, A.H.C. (promotor), Olabarriaga, S. (co-promotor). [ UvA-DARE ]
- Anang, D. (2023) 6 November 2023. B and T Cell Immune Responses in Rheumatoid Arthritis and Myositis. In Search for the Immunological Drummers and Dancers . University of Amsterdam, Amsterdam. COSMIC . Marie Curie ITN, Horizon 2020. De Vries, N. (promotor), Van Kampen, A.H.C. (promotor), van Baarsen, E.G.M. (co-promotor). [ UvA-DARE ]
- Wegdam, W. (2024). In search of protein biomarkers in ovarian cancer and Gaucher disease. University of Amsterdam, Amsterdam. Aerts J.M.F.G. (promotor), Kenter, G.G. (promotor), Moerland, P.D. (co-promotor). [ UvA-DARE ]
- Pollastro, S (2024) 17 May 2024. Understanding Response to Rituximab Treatment in Rheumatoid Arthritis Through Immune Fingerprinting of T and B Cells . University of Amsterdam, Amsterdam. De Vries, N. (promotor), Van Kampen, A.H.C. (co-promotor). [ UvA-DARE ].

Related Posts

PhD thesis Umesh Nandal
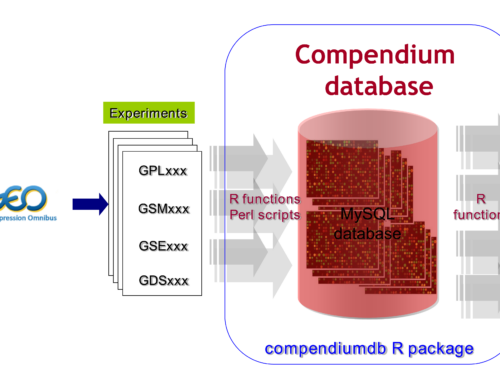
compendiumdb

Participation in single-cell genomics 2022 conference

PhD thesis Elena Merino Tejero
Group Leader
Prof. dr. AHC van Kampen
[email protected] https://bioinformaticslaboratory.eu

Amsterdam UMC – location AMC Department of Epidemiology and Data Science Bioinformatics Laboratory Meibergdreef 9 1105 AZ Amsterdam Zuidoost The Netherlands
How to get to us: [ pdf ] [ Google Maps ]
Epidemiology & Data Science
The Bioinformatics Laboratory is part of EDS

Read our Privacy Policy
Bioinformatics Laboratory – Your partner in bioinformatics and computational modelling since 1997
Privacy Overview
| Cookie | Duration | Description |
|---|---|---|
| cookielawinfo-checkbox-analytics | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Analytics". |
| cookielawinfo-checkbox-functional | 11 months | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Functional". |
| cookielawinfo-checkbox-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-others | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Other. |
| cookielawinfo-checkbox-performance | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Performance". |
| viewed_cookie_policy | 11 months | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |
BSc and MSc Thesis Subjects of the Bioinformatics Group
On this page you can find an overview of the BSc and MSc thesis topics that are offered by our group. The procedure to find the right thesis project for you is described below.
MSc thesis: In the Bioinformatics group, we offer a wide range of MSc thesis projects, from applied bioinformatics to computational method development. Here is a list of available MSc thesis projects . Besides the fact that these topics can be pursued for a MSc thesis, they can also be pursued as part of a Research Practice .
BSc thesis: As a BSc student you will work as an apprentice alongside one of the PhD students or postdocs in the group. You will work on your own research project, closely guided by your supervisor. You will be expected to work with several tools and/or databases, be creative and potentially overcome technical challenges. Below you will find short descriptions of the research projects of our PhDs and Postdocs. In addition you can take a look at the list of MSc thesis projects above.
Procedure for WUR students:
- Request an intake meeting with one of our thesis coordinators by filling out the MSc intake form or BSc intake form and sending it to [email protected]
- Contact project supervisors to discuss specific projects that fit your background and interest
- Upon a match, take care of the required thesis administration together with your supervisor(s) and enroll in the thesis BrightSpace site to find more information on a thesis in the Bioinformatics group
Procedure for non-WUR students or students in other non-standard situations: We have limited space for interns from other institutes. If you are interested, please email our thesis coordinators at [email protected]; please attach your CV and indicate what are your main research interests.
BSc thesis topics
Integrative omics for the discovery of biosynthetic pathways in plants, molecular function prediction of natural products, linking the metabolome and genome, linking metagenomics and metatranscriptomics to study the endophytic root microbiome, exploiting variation in lettuce and its wild relatives.
- Fellowships and Financial Aid
- Frequently Asked Questions
- Masters in Bioinformatics
- Administration
- NIH Training Grant
- Student Support Services
- Commitment to Inclusion
- Volunteering and Outreach
- Learning Outcomes
- PhD & MS Resources
- Course Schedule
- Computational Resources
- Bioinformatics Program Retreat
- Student-Organized Symposium
- International Workshop on Bioinformatics and Systems Biology
PhD Thesis Defenses
January 4th Kritika Karri
Title: Computational Characterization of Long Non-Coding RNAs (LncRNAs) and Study Their Role in Rodent Liver Disease, Xenobiotic Exposure, and Sex-Specific Responses Using Bulk and Single Cell RNA-Sequencing
Major Professor: David Waxman
ABSTRACT: LncRNAs comprise a heterogeneous class of thousands of RNA-encoding genes whose functions are largely unknown. This thesis describes systematic computational approaches to discover liver-expressed lncRNAs globally and then deduce their regulatory roles in response to foreign chemical and hormonal exposures. In a first study, bulk liver RNA-seq data was used to discover liver-expressed lncRNAs responsive to multiple xenobiotics in a rat model. Ortholog analysis combined with co-expression data and causal inference methods was used to infer lncRNA function and deduce gene regulatory networks, including causal effects of lncRNAs on biological pathways. This work provides a framework for understanding the widespread transcriptome-altering actions of foreign chemicals in a key-responsive mammalian tissue. In a second study, single-cell RNA-seq was employed to develop a reference catalog of 48,261 mouse liver-expressed lncRNAs, a majority novel, by transcriptome reconstruction from > 2,000 bulk public mouse liver RNA-seq datasets. Single cell RNA-seq was sufficiently sensitive to detect >30,000 mouse liver lncRNAs and characterize their dysregulation in mouse models of high fat diet-induced non-alcoholic steatohepatitis (NASH), carbon tetrachloride-induced liver fibrosis, and hepatotoxicity induced by the Ah receptor agonist TCDD. Trajectory inference algorithms uncovered lncRNA zonation patterns in five major hepatic cell populations and their dysregulation in diseased states. LncRNAs expressed in NASH-associated macrophages, closely linked to disease progression, and in collagen-producing myofibroblasts, a key source of the fibrous scar in fibrotic liver, were identified. Regulatory network analysis linked individual lncRNAs with key biological pathways and gene centrality metrics identified network-essential regulatory lncRNAs in each liver disease model. In a third study, single nucleus RNA-seq combined with single nucleus ATAC-seq mapping of open chromatin regions elucidated functional linkages between cis- and trans-regulatory elements and their downstream genes targets, notably genes showing expression sex-differences impacting metabolism and disease risk. Liver cell type-specific chromatin accessibility signatures were identified, as were sex-specific accessibility signatures for hepatocytes and their associated DNA regulatory region motifs. Integrative modalities were employed to elucidate transcription factor-based mechanisms involved in sex-specific growth hormone-regulated gene expression by identifying transcriptional and epigenetic changes during feminization of mouse liver. Together, these studies characterize lncRNA function and can motivate future experiments.
June 10 Aaron Chevalier
Title: Tools for Mutational Signature Discovery and Methods for Prediction of Drug Response
Major Professor: Joshua Campbell
ABSTRACT: Mutational signatures are patterns of somatic alterations in the genome caused by carcinogenic exposures or aberrant cellular processes. Specifically, this dissertation focuses on the analysis of mutational signatures in human cancer and its application to stratification of patients for drug response.
To provide a comprehensive workflow for preprocessing, analysis, and visualization of mutational signatures, I created the Mutational Signature Comprehensive Analysis Toolkit (musicatk) package. musicatk enables users to select different schemas for counting mutation types and easily combine count tables from different schemas. Multiple distinct methods are available to deconvolute signatures and exposures or to predict exposures in individual samples given a pre-existing set of signatures. Additional exploratory features include the ability to compare signatures to the COSMIC database, embed tumors in two dimensions with UMAP, cluster tumors into subgroups based on exposure frequencies, identify differentially active exposures between tumor subgroups, and plot exposure distributions across user-defined annotations such as tumor type.
I then use musicatk to analyze the largest tumor sequencing dataset from a Chinese population to date. I identified differences in the levels of signature exposures compared to similar data from a Western cohort. Specifically, COSMIC signature SBS25 was higher in the Chinese dataset for Melanoma and Renal Cell Carcinoma patients and Melanoma patients had lower levels of SBS7a/b (Ultraviolet Light). My analysis also revealed a putative novel signature enriched in pancreatic cancers.
Lastly, I assess the ability of mutational signatures to identify patients who may respond to irofulven, a drug for late-stage cancer patients who have defects in the Transcription Coupled Nucleotide Excision Repair (TC-NER) pathway. As the functional understanding of which mutations successfully disrupt this pathway is incomplete, I develop an approach that classifies patients based on evidence of this pathway being disrupted based on levels of mutational signatures. I build a model that successfully predicts patients who will respond to treatment without a known relevant mutation in the TC-NER pathway.
The work from this study furthers our understanding of mutational signatures in different populations and demonstrates the feasibility of using mutational signatures to identify patients eligible for drug trials.
August 19 Lucas Schiffer
Title: Multimodal, Longitudinal, and Mega-Analysis of Biomedical Data
Major Professor: W. Evan Johnson
ABSTRACT: Biomedical data science is a multi-disciplinary field concerned with the collection, storage, and interpretation of biomedical data that uses annotation, algorithms, and analysis to extract knowledge and insights from structured and unstructured data to be used in the development and evaluation of diagnostic tests, prognostic predictions, and therapeutic interventions. Biomedical data scientists perform this work using biomedical data that arises when samples are subjected to biochemical assays to quantitively or qualitatively investigate their pathophysiological characteristics. Increasingly, biomedical data are generated at single-cell resolution and have consequently become far more hierarchical and multimodal in nature – that is, levels of organization encapsulate one another (e.g., samples belonging to subjects are made up of cells) and multiple biological modalities are profiled simultaneously. The paradigm shift adds significant complexity to the collection, storage, management, and analysis of biomedical data, but brings with it the promise of unprecedent insights to be gained from integrative analyses. These analyses are the focus of this dissertation, where the challenges of integrating biomedical data across multiple modalities, timepoints, and studies are examined through three research projects.
Challenges related to multimodal analysis of biomedical data will be explored through the development of MultimodalExperiment, a data structure that appropriately and efficiently represents multiomics data that is hierarchical, multimodal, and/or longitudinal in nature. A schematic of and methods for the data structure will be presented along with example usage to demonstrate how current challenges of alternative data structures are overcome, ease of data management is improved, and computational/storage efficiency is optimized.
Challenges related to longitudinal analysis of biomedical data will be explored in the context of a cohort study of cancer patients being treated with anti-programmed cell death protein 1/programmed cell death ligand 1 immunotherapies at Boston Medical Center. The progression-free survival status of study participants will be analyzed using linear mixed effects models which incorporate longitudinal high-dimensional metabolomics data. Maps of metabolic pathways and a hypothesis will be presented to explain serum metabolites that are associated with progress-free survival status and possibly therapeutic efficacy.
Challenges related to mega-analysis of biomedical data will be explored through the creation of a pipeline to preprocess transcriptomics data from human host infected with tuberculosis to support machine learning and other tasks. The details of original software developed to provide more than 10,000 samples of clean high-quality machine learning ready data from all related and eligible studies in the Gene Expression Omnibus repository will be illustrated. The importance improving diagnostic testing and therapeutic interventions for tuberculosis disease will be highlighted in the context of these data, and the specifics of why they represent a key ingredient for machine learning that helps overcome current challenges in the field will be explained.
August 24 Boting Ning
Title: Leveraging Transcriptomic Regulation to Understand, Diagnose and Intercept Early Lung Cancer Pathogenesis
Major Professor: Marc Lenburg
ABSTRACT: Lung cancer is the leading cause of cancer death in the US, largely due to the lack of treatment options to intercept the progression of early lung cancers and methods to diagnose lung cancer at early stages. Prior studies indicated that the lack of immune surveillance is associated with the progression of bronchial premalignant lesions (PMLs) and the gene alterations in the nasal epithelium can be leveraged for the early detection of lung cancer. Yet, the regulatory mechanism of these gene expression alterations is still less understood. Thus, there are unmet needs to study the gene expression regulation for better disease management of early lung cancer, including further understanding the biology of early lung cancer development, identifying potential interception strategies, and improving the lung cancer diagnosis.
My dissertation addresses these challenges by investigating the transcriptional and post-transcriptional gene expression regulators, including transcription factors and microRNAs (miRNAs), to facilitate the understanding, interception, and diagnosis of early lung cancer. First, I explored the miRNA regulatory landscape to identify miRNA-gene regulatory relationships associated with bronchial PML progression and molecular subtypes. Using matched gene and microRNA expression profiles from patients with bronchial premalignant lesions, I identified epithelial miR-149-5p to be a key regulator of gene expression contributing to PML progression. By suppressing NLRC5, miR-149-5p inhibits MHC-I gene expression of epithelial cells, promoting early immune depletion and lesion progression. I also developed a novel statistical framework, Differential Regulation Analysis of miRNA (DReAmiR), that characterizes miRNA-mediated gene regulatory network rewiring across multiple groups from transcriptomic profiles, and identified regulatory network differences across PML molecular subtypes. Secondly, I investigated the alterations in the Hippo pathway to identify potential drug targets to intercept the progression of bronchial PMLs. I found that Hippo pathway effectors YAP/TAZ, together with transcription factors TEAD and TP63, cooperatively promote basal cell proliferation and repress signals associated with interferon responses and immune cell communication. Further in silico drug screening with external datasets identified small compounds that can reverse the direct regulated gene signature to potentially intercept bronchial PML progression. Lastly, I integrated miRNA and gene expression profiles in the nasal epithelium to distinguish malignant from benign indeterminate pulmonary nodules. I built an ensemble classifier consisting of nasal epithelial miRNA expression features, miRNA-gene top scoring pairs, and clinical features. The performance of the ensemble classifier exceeded that of the classifier built with clinical features alone.
Collectively, my thesis investigated the gene expression regulation mechanisms to facilitate the understanding, interception, and diagnosis of early lung cancer pathogenesis.
November 17th Rebecca Panitch
Title: Understanding the Mechanisms and Pathways of Alzheimer’s Disease in APOE Genotype Sub-Populations
Major Professor: Lindsay Farrer
ABSTRACT: Alzheimer’s disease (AD) is a neurodegenerative disease classified pathologically by the presence of tau tangles and amyloid plaques. The largest genetic risk factor for AD is the APOE ε4 allele, while the APOE ε2 allele has been linked to a protective effect for AD. Recent studies demonstrated that APOE genotypes are linked to unique omics signatures and pathological features relating to AD, such as blood-brain barrier breakage. To investigate the role of APOE genotype in AD, I analyzed different levels of omic data in blood and brain. I analyzed transcriptomic data derived from autopsied brains using network and differential gene expression approaches to identify genes and pathways involved in the APOE ε2 protective mechanism for AD. Additionally, I identified APOE genotype-specific pathways and networks involved in both blood and brain function in AD using blood and brain tissue gene expression from mostly the same individuals. Lastly, I analyzed the association of methylation of DNA from blood and brain samples with AD to identify APOE and AD specific methylation signatures and potential drug targets. Collectively, this thesis emphasizes the utility of investigating APOE genotypes individually to identify novel pathways and potential drug targets within AD subpopulations.
November 21st Dileep Kishore
Title: Computational Study of Microbe-Microbe Interactions and Their Interplay with Their Environment
Major Professor: Daniel Segrè
ABSTRACT: Microbial communities play important roles in human health and disease, are essential components of terrestrial and marine ecosystems, and are crucial for producing commercially valuable molecules in industrial processes. These communities consist of hundreds of species involved in complex interactions. Mapping the interrelationships between different species in a microbial community is vital for understanding and controlling ecosystem structure and function. Advances in sequencing and other omics technologies have led to thousands of datasets containing information about microbial composition, gene expression, and metabolism in microbial communities associated with human hosts and other environments. These provide valuable information in understanding how microbes interact with each other and how their interactions affect the health of their host (e.g., human or plant). Furthermore, understanding these interactions paves the way for the rational design and modulation of synthetic communities for producing antibiotics, biofuels, and pharmaceutical products.
The first part of my thesis is focused on improving the workflow for the inference of microbial co-occurrence relationships from abundance data. Toward this goal, we developed Microbial Co-occurrence Network Explorer or MiCoNE, a pipeline that infers microbial co-occurrences from 16S ribosomal RNA (16S rRNA) amplicon data. The second part of my thesis focuses on microbe-host interactions rather than microbe-microbe associations. In particular, we sought to predict the effects of microbial metabolites on human receptors and their associated regulatory pathways. In the final part of my thesis, we turn to the question of whether computational algorithms can help control microbial community growth to achieve specific objectives. We describe the development of a reinforcement learning algorithm to learn optimal environmental control strategies to steer a microbial community towards a particular goal, such as reaching a specific taxonomic distribution or producing desired metabolites.
Overall, the work presented in this thesis demonstrates how microbe-microbe and microbe-environment (including microbe-host) interactions represent plastic system-level properties whose understanding can help unravel the role of microbial communities in specific diseases. Correspondingly, manipulating these interactions, e.g., by appropriately modifying environmental conditions, can serve as a promising strategy for steering communities towards desired states, including producing valuable molecular products.
December 9th Rui Hong
Title: Building an Analytical Framework for Quality Control and Meta-Analysis of Single-Cell Data to Understand Heterogeneity in Lung Cancer Cells
ABSTRACT: Single-cell RNA sequencing (scRNA-seq) has been a powerful technique for characterizing transcriptional heterogeneity related to tumor development and disease pathogenesis. Despite the advances of the technology, there is still a lack of software to systematically and easily assess the quality and different types of artifacts present in scRNA-seq data and lack of statistical frameworks for understanding heterogeneity in the gene programs of cancer cells.
In this dissertation, I first introduced novel computational software to enhance and streamline the process of quality control for scRNA-seq data called SCTKQC. SCTK-QC is a pipeline that performs comprehensive quality control (QC) of scRNA-seq data and runs a multitude of tools to assess various types of noise present in scRNA-seq data as well as quantification of general QC metrics. These metrics are displayed in an user-friendly HTML report and the pipeline has been implemented in two cloud-based platforms.
Most scRNA-seq studies only profiled a small number of tumors and provided a narrow view of the transcriptome in tumor tissue. Next, I developed a novel framework to perform a large-scale meta-analysis of cancer cells from 12 studies with scRNA-seq data from patients with non-small-cell lung cancer (NSCLC). I discovered interpretable gene co-expression modules with celda and demonstrated that the activity of gene modules accounted for both inter- and intra-tumor heterogeneity of NSCLC samples. Furthermore, I used CaDRa to determine that the levels of some gene modules were significantly associated with combinations of underlying genetic alterations. I also show that other gene modules are associated with immune cell signatures and may be important for communication with the cancer cells and the immune microenvironment.
Finally, I presented a novel computational method to study the association between copy number variation (CNV) and gene expression at single-cell level. The diversity of CNV profile was identified in tumor subclones within each sample and I discovered cis and trans gene signatures which have expression value associated with specific somatic CNV status. This study helped us prioritize the potential cancer driver genes within each CNV region.
Collectively, this work addressed the limitation in the quality control of scRNAseq data and provided insights for understanding the heterogeneity of NSCLC samples.
December 2 Emma Briars
Title: Development Of Methods To Diagnose And Predict Antibiotic Resistance Using Synthetic Biology And Computational Approaches
Major Professor: Ahmad (Mo) Khalil
ABSTRACT: Antibiotic resistance is a quickly emerging public health crisis, accounting for more than 700,000 annual global deaths. Global human antibiotic overuse and misuse has significantly expedited the rate at which bacteria become resistant to antibiotics. A renewed focus on discovering new antibiotics is one approach to addressing this crisis. However, it alone cannot solve the problem: historically, the introduction of a new antibiotic has consistently, and at times rapidly, been followed by the appearance and dissemination of resistant bacteria. It is thus crucial to develop strategies to improve how we select and deploy antibiotics so that we can control and prevent the emergence and transmission of antibiotic resistance. Current gold-standard antibiotic susceptibility tests measure bacterial growth, which can take up to 72 hours. However, bacteria exhibit more immediate measurable phenotypes of antibiotic susceptibility, including changes in transcription, after brief antibiotic exposure. In this dissertation I develop a framework for building a paper-based cell-free toehold sensor antibiotic susceptibility test that can detect differential mRNA expression. I also explore how long-term lab evolution experiments can be used to prospectively uncover transcriptional signatures of antibiotic susceptibility.
Paper-based cell-free systems provide an opportunity for developing clinically tractable nucleic-acid based diagnostics that are low-cost, rapid, and sensitive. I develop a computational workflow to rapidly and easily design toehold switch sensors, amplification primers, and synthetic RNAs. I develop an experimental workflow, based on existing paper-based cell-free technology, for screening toehold sensors, amplifying bacterial mRNA, and deploying sensors for differential mRNA detection. I combine this work to introduce a paper-based cell-free toehold sensor antibiotic susceptibility test that can detect fluoroquinolone-susceptible E. coli. Next, I describe a methodology for long-term lab evolution and how it can be used to explore the relationship between a phenotype, such as gene expression, and antibiotic resistance acquisition. Using a set of E. coli strains evolved to acquire tetracycline resistance, I explore how each strains transcriptome changes as resistance increases. Together, this work provides a set of computational and experimental methods that can be used to study the emergence of antibiotic resistance, and improve upon available methods for properly selecting and deploying antibiotics.
November 18 Anthony Federico
Title: Development of Methods for Omics Network Inference and Analysis and Their Application to Disease Modeling
Major Professor: Stefano Monti
ABSTRACT: With the advent of Next Generation Sequencing (NGS) technologies and the emergence of large publicly available genomics data comes an unprecedented opportunity to model biological networks through a holistic lens using a systems-based approach. Networks provide a mathematical framework for representing biological phenomena that go beyond standard one-gene-at-a-time analyses. Networks can model system-level patterns and the molecular rewiring (i.e., changes in connectivity) occurring in response to perturbations or between distinct phenotypic groups or cell types. This in turn supports the identification of putative mechanisms of actions of the biological processes under study, and thus has the potential to advance prevention and therapy. However, there are major challenges faced by researchers. Inference of biological network structures is often performed on high-dimensional data, yet is hindered by the limited sample size of high throughput omics data. Furthermore, modeling biological networks involves complex analyses capable of integrating multiple sources of omics layers and summarizing large amounts of information.
My dissertation aims to address these challenges by presenting new approaches for high-dimensional network inference with limited samples as well as methods and tools for integrated network analysis applied to multiple research domains in cancer genomics. First, I introduce a novel method for reconstructing gene regulatory networks called SHINE (Structure Learning for Hierarchical Networks) and present an evaluation on simulated and real datasets including a Pan-Cancer analysis using The Cancer Genome Atlas (TCGA) data. Next, I summarize the challenges with executing and managing data processing workflows for large omics datasets on high performance computing environments and present multiple strategies for using Nextflow for reproducible scientific workflows including shine-nf – a collection of Nextflow modules for structure learning. Lastly, I introduce the methods, objects, and tools developed for the analysis of biological networks used throughout my dissertation work. Together – these contributions were used in focused analyses of understanding the molecular mechanisms of tumor maintenance and progression in subtype networks of Breast Cancer and Head and Neck Squamous Cell Carcinoma.
August 4 Brian Haas
Title: Bioinformatic Tool Developments with Applications to RNA-Seq Data Analysis and Clinical Cancer Research
Major Professors: Simon Kasif & Aviv Regev
ABSTRACT Modern advances in sequencing technologies have enabled exploration of molecular biology at unprecedented scale and resolution. Transcriptome sequencing (RNA-seq), in particular, has been widely adopted as a routine cost-effective method for assaying both genetic and functional characteristics of biological systems with resolution down to individual cells. Clinical research and applications leveraging these technologies have largely targeted tumor biology, where transcriptome sequencing can capture tumor genetic and epigenetic characteristics and aid with understanding the etiology or guide treatments. Specialized computational methods and bioinformatic software tools are essential for processing and analyzing RNA-seq to explore various aspects of tumor biology including driver mutations, genome rearrangements, and aneuploidy. With single cell resolution, such methods can yield insights into tumor cellular composition and heterogeneity. Here, we developed methods and tools to support cancer transcriptome studies for bulk and single cell tumor transcriptomes, focusing primarily on fusion transcript detection and predicting large-scale copy number alternations from RNA-seq. These efforts culminated in the development of STAR-Fusion for fast and accurate detection of fusion transcript, FusionInspector for further characterizing predicted fusion transcripts and discriminating likely artifacts, and TrinityFusion for de novo reconstruction of fusion transcripts and tumor viruses. We also developed advanced methods for predicting copy number alterations and subclonal architecture from tumor and normal single cell RNA-seq data, as incorporated into our InferCNV software. In addition to these bioinformatic method and software development, we applied our fusion detection methods to thousands of tumor and normal samples and gain novel insights that should further help guide researchers with clinical applications of fusion transcript discovery.
July 29 Tanya Karagiannis
Title: Single Cell Analysis and Methods To Characterize Peripheral Blood Immune Cell Types in Disease and Aging
Major Professors: Stefano Monti & Paola Sebastiani
ABSTRACT In the past decade, RNA-sequencing (RNA-seq)-based genome-wide expression studies have contributed to major advances in understanding human biology and disease. However, for heterogeneous tissues such as peripheral blood, RNA-sequencing masks the expression of different populations of cells that may be important in understanding different conditions and disease progression. With the advent of single cell RNA-sequencing (scRNA-seq), it has become possible to study the gene expression of each single cell and to explore cellular heterogeneity in the context of disease and under the influence of medications or other substances. In this dissertation, I will present three projects that demonstrate how single cell sequencing methods can be used to characterize novel changes in the peripheral immune system in human disease and aging. I will also describe novel methodological approaches I created to analyze cell type composition and gene expression level changes.
First, I investigated the cell type specific changes due to opioid use in human peripheral blood. Utilizing single cell transcriptomic methods, I identified a genome-wide suppression of antiviral gene expression across immune cell types of chronic opioid users, and similarly under acute exposure to morphine.
Second, I investigated the immune cell type specific changes of gene expression and composition in the context of human aging and longevity. I developed novel approaches to measure and compare overall cell type composition between samples, and identified significant overall differences in immune cell type composition, including pro-inflammatory cell populations, between extreme longevity and younger ages. In addition, I generated cell type-specific signatures associated with longevity after accounting for age-related changes that demonstrate an upregulation in immune response and metabolic processes important in the activation of immune cells in extreme long-lived individuals compared to normally aging individuals.
Finally, I investigated whether aging of the immune system is accelerated in opioid-dependent individuals. I utilized the unique aging signatures generated in the aging project and discovered higher expression of aging signatures in specific cell types of opioid-dependent individuals, suggesting chronic opioid use causes premature aging of the immune system that may contribute to the increased susceptibility to infections in these individuals.
March 24th Marzie Rasekh
Title: Characterizing VNTRS in Human Populations
Major Professor: Gary Benson
ABSTRACT Over half the human genome consists of repetitive sequences. One major class is the tandem repeats (TRs), which are defined by their location in the genome, repeat unit, and copy number. TRs loci which exhibit variant copy numbers are called Variable Number Tandem Repeats (VNTRs). High VNTR mutation rates of approximately 10-4 per generation make them suitable for forensic studies, and of interest for potential roles in gene regulation and disease. TRs are generally divided into three classes: 1) microsatellites or short tandem repeats (STRs) with patterns <7 bp; 2) minisatellites with patterns of seven to hundreds of base pairs; and 3) macrosatellites with patterns of >100 bp. To date, mini- and macrosatellites have been poorly characterized, mainly due to a lack of computational tools. In this thesis, I utilize a tool, VNTRseek, to identify human minisatellite VNTRs using short read sequencing data from nearly 2,800 individuals and developed a new computational tool, MaSUD, to identify human macrosatellite VNTRs using data from 2,504 individuals. MaSUD is the first high-throughput tool to genotype macrosatellites using short reads.
I identified over 35,000 minisatellite VNTRs and over 4,000 macrosatellite VNTRs, most previously unknown. A small subset in each VNTR class was validated experimentally and in silico. The detected VNTRs were further studied for their effects on gene expression, ability to distinguish human populations, and functional enrichment. Unlike STRs, mini- and macrosatellite VNTRs are enriched in regions with functional importance, e.g., introns, promoters, and transcription factor binding sites. A study of VNTRs across 26 populations shows that minisatellite VNTR genotypescan be used to predict super-populations with >90% accuracy. In addition, genotypes for 195 minisatellite VNTRs and 24 macrosatellite VNTRs were shown to be associated with differential expression in nearby genes (eQTLs).
Finally, I developed a computational tool, mlZ, to infer undetected VNTR alleles and to detect false positive predictions. mlZ is applicable to other tools that use read support for predicting short variants.
Overall, these studies provide the most comprehensive analysis of mini- and macrosatellites in human populations and will facilitate the application of VNTRs for clinical purposes.
April 8th Zhe Wang
Title: Enhancing Preprocessing and Clustering of Single-Cell RNA Sequencing Data
ABSTRACT Single-cell RNA sequencing (scRNA-seq) is the leading technique for characterizing cellular heterogeneity in biological samples. Various scRNA-seq protocols have been developed that can measure the transcriptome from thousands of cells in a single experiment. With these methods readily available, the ability to transform raw data into biological understanding of complex systems is now a rate-limiting step. In this dissertation, I introduce novel computational software and tools which enhance preprocessing and clustering of scRNA-seq data and evaluate their performance compared to existing methods.
First, I present scruff, an R/Bioconductor package that preprocesses data generated from scRNA-seq protocols including CEL-Seq or CEL-Seq2 and reports comprehensive data quality metrics and visualizations. scruff rapidly demultiplexes, aligns, and counts the reads mapped to genomic features with deduplication of unique molecular identifier (UMI) tags and provides novel and extensive functions to visualize both pre- and post-alignment data quality metrics for cells from multiple experiments.
Second, I present Celda, a novel Bayesian hierarchical model that can perform simultaneous co-clustering of genes into transcriptional modules and cells into subpopulations for scRNA-seq data. Celda identified novel cell subpopulations in a publicly available peripheral blood mononuclear cell (PBMC) dataset and outperformed a PCA-based approach for gene clustering on simulated data.
Third, I extend the application of Celda by developing a multimodal clustering method that utilizes both mRNA and protein expression information generated from single-cell sequencing datasets with multiple modalities, and demonstrate that Celda multimodal clustering captured meaningful biological patterns which are missed by transcriptome- or protein-only clustering methods.
Collectively, this work addresses limitations present in the computational analyses of scRNA-seq data by providing novel methods and solutions that enhance scRNA-seq data preprocessing and clustering.
April 8th Ke Xu
Title: Airway Gene Expression Alterations in Association with Radiographic Abnormalities of the Lung
ABSTRACT High-resolution computed tomography (HRCT) of the chest is commonly used in the diagnosis of a variety of lung diseases. Structural changes associated with clinical characteristics of disease may also define specific disease-associated physiologic states that may provide insights into disease pathophysiology. Gene expression profiling is potentially a useful adjunct to HRCT to identify molecular correlates of the observed structural changes. However, it is difficult to directly access diseased distal airway or lung parenchyma routinely for profiling studies.
Previously, we have profiled bronchial airway in normal-appearing epithelial cells at the mainstem bronchus, detecting distinct gene expression alterations related to the clinical diagnosis of chronic obstructive pulmonary disease (COPD) and lung cancer. These gene expression alterations offer insights into the molecular events related to diseased tissue at more distal airways and in the parenchyma, which we hypothesize are due to a field-of-injury effect. Here, we expand this prior work by correlating airway gene expression to COPD and bronchiectasis phenotypes defined by HRCT to better understand the pathophysiology of these diseases. Additionally, we classified pulmonary nodules as malignant or benign by combining HRCT nodule imaging characteristics with gene expression profiling of the nasal airway.
First, we collected brushing samples from the main-stem bronchus and assessed gene expression alterations associated with COPD phenotypes defined by K-means clustering of HRCT-based imaging features. We found three imaging clusters, which correlated with incremental severity of COPD: normal, interstitial predominant, and emphysema predominant. 41 genes were differentially expressed between the normal and the emphysema predominant clusters. Functional analysis of the differentially expressed genes suggests a possible induction of inflammatory processes and repression of T-cell related biologic pathways, in the emphysema predominant cluster.
We then discovered gene expression alterations associated with radiographic evidence of bronchiectasis (BE), an underdiagnosed obstructive pulmonary disease with unclear pathophysiology. We found 655 genes were differentially expressed in bronchial epithelium from individuals with radiographic evidence of BE despite none of the study participants having a clinical BE diagnosis. In addition to biological pathways that had been previously associated with BE, novel pathways that may play important roles in BE initiation were also discovered. Furthermore, we leveraged an independent single-cell RNA-sequencing dataset of the bronchial epithelium to explore whether the observed gene expression alterations might be cell-type dependent. We computationally detected an increased presence of ciliated and deuterosomal cells, as well as a decreased presence of basal cells in subjects with widespread radiographic BE, which may reflect a shift in the cellular landscape of the airway during BE initiation.
Finally, we identified gene expression alterations within the nasal epithelium associated with the presence of malignant pulmonary nodules. A computational model was constructed for determining whether a nodule is malignant or benign that combines gene expression and imaging features extracted from HRCT. Leveraging data from single-cell RNA sequencing, we found genes increased in patients with lung cancer are expressed at higher levels within a novel cluster of nasal epithelial cells, termed keratinizing epithelial cells.
In summary, we leveraged gene expression profiling of the proximal airway and discovered novel biological pathways that potentially drive the structural changes representative of physiologic states defined by chest HRCT in COPD and BE. This approach may also be combined with chest HRCT to detect weak signals related to malignant pulmonary nodules.
December 3rd Tyler Faits
Title: The Evaluation, Application, and Expansion of 16S Amplicon Metagenomics
ABSTRACT Since the invention of high-throughput sequencing, the majority of experiments studying bacterial microbiomes have relied on the PCR amplification of all or part of the gene for the 16S rRNA subunit, which serves as a biomarker for identifying and quantifying the various taxa present in a microbiomic sample. Several computational methods exist for analyzing 16S amplicon based metagenomics, but the most commonly used bioinformatics tools are unable to produce quality genus-level or species-level taxonomic calls and may underestimate the degree to which such calls are possible. In this thesis, I have used 16S sequencing data from mock bacterial communities to evaluate the sensitivity and specificity of several bioinformatics pipelines and genomic reference libraries used for microbiome analyses, with a focus on measuring the accuracy of species-level taxonomic assignments of 16S amplicon reads. With the efficacy of these tools established, I then applied them in the analysis of data from two studies into human microbiomes. I evaluated the metagenomics analysis tools Qiime 2, Mothur, PathoScope 2, and Kraken, in conjunction with reference libraries from GreenGenes, Silva, Kraken, and RefSeq, using publicly available mock community data from several sources, comprising 137 samples spanning a range of taxonomic diversity, amplicon regions, and sequencing methods. PathoScope and Kraken, both tools designed for whole genome metagenomics, outperformed Qiime 2 and Mothur, which are theoretically specialized in 16S analyses. I used PathoScope 2 to analyze longitudinal 16S data from infants in Zambia, exploring the maturation of nasopharyngeal microbiomes in healthy infants, establishing a range of typical healthy taxonomic profiles, and identifying dysbiotic patterns which are associated with the development of severe lower respiratory tract infections in early childhood. With more data, these dysbiotic patterns may help identify infants at high risk of developing respiratory disease.
I used Qiime 2 to analyze 16S data from human subjects in a controlled dietary intervention study with a focus on dietary carbohydrate quality. I correlated alterations in the gut microbiome with various cardiometabolic risk factors, and identified increases in some butyrate-producing bacteria in response to complex carbohydrates. I also constructed a metatranscriptomics pipeline to analyze paired rRNA-depleted RNAseq data.
October 14th Alan Pacheco
Title: Environmental Modulation of Microbial Ecosystems
Major Professor: Daniel Segre
ABSTRACT Natural microbiota are essential to the health of living systems – from the human gut to coral reefs. Although advances in DNA sequencing have allowed us to catalogue many of the different organisms that make up these microbial communities, significant challenges remain in understanding the complex networks of interspecies metabolic interactions they exhibit. These interactions are crucial to community stability and function, and are highly context-dependent: the availability of different nutrients can determine whether a set of microbes will interact cooperatively or competitively, which can drastically change a community’s structure. Disentangling the environmental factors that determine these behaviors will not only fundamentally enhance our knowledge of their ecological properties, but will also bring us closer to the rational engineering of synthetic microbiomes with novel functions. Here, I integrate modeling and experimental approaches to quantify the dependence of microbial communities on environmental composition. I then show how this relationship can be leveraged to facilitate the design of synthetic consortia.
The first chapter of this dissertation is a review article that introduces a framework for cataloguing interaction mechanisms, which enables quantitative comparisons and predictive models of these complex phenomena. The second chapter is a computational study that explores one such attribute – metabolic cost – in high detail. It demonstrates how a large variety of molecules can be secreted without imposing a fitness cost on microbial organisms, allowing for the emergence of beneficial interspecies interactions. The third chapter is an experimental study that determines how the number of unique environmental nutrients affects microbial community growth and taxonomic diversity. The integration of stoichiometric and consumer resource models enabled the discovery of basic ecological principles that govern this environment phenotype relationship. The fourth chapter applies these principles to the design of engineered communities via a search algorithm that identifies environmental compositions that yield specific ecosystem properties. This dissertation then concludes with extensions of the modeling methods used throughout this work to additional model systems.
Future work could further quantify how microbial community phenotypes depend on each of the individual factors explored in this thesis, while also leveraging emerging knowledge on interaction mechanisms to design synthetic consortia.
August 24th Devanshi Patel
Title: Tissue-Dependent Analysis of Common and Rare Genetic Variants for Alzheimer’s Disease Using Multi-Omics Data
ABSTRACT Alzheimer’s disease (AD) is a complex neurodegenerative disease characterized by progressive memory loss and caused by a combination of genetic, environmental, and lifestyle factors. AD susceptibility is highly heritable at 58-79%, but only about one third of the AD genetic component is accounted for by common variants discovered through genome-wide association studies (GWAS). Rare variants may contribute to some of the unexplained heritability of AD and have been demonstrated to contribute to large gene expression changes across tissues, but conventional analytical approaches pose challenges because of low statistical power even for large sample sizes. Recent studies have demonstrated by expression quantitative trait locus (eQTL) analysis that changes in gene expression could play a key role in the pathogenesis of AD. However, regulation of gene expression has been shown to be context-specific (e.g., tissue and cell-types), motivating a context dependent approach to achieve more precise and statistically significant associations. To address these issues, I applied a strategy to identify new AD risk or protective rare variants by examining mutations occurring only in cases or only controls, observing that different mutations in the same gene or variable dose of a mutation may result in distinct dementias. I also evaluated the impact of rare variation on expression at the gene and gene pathway levels in blood and brain tissue, further strengthening the rare variant findings with functional evidence and finding evidence for a large immune and inflammatory component to AD. Lastly, I identified cell-type specific eQTLs in blood and brain tissue to explain underlying genetic associations of common variants in AD, and also discovered additional evidence for the role of myeloid cells in AD risk and potential novel blood and brain AD biomarkers. Collectively, these findings further explain the genetic basis of AD risk and provide insight about mechanisms leading to this disorder.

- Research Partnerships
Prospective Students
- Degree Programs
Seminars & Events
- 7 / 18 BU Microbiome Day
- 7 / 18 Bioinformatics Challenge Project Presentations
- Mobile Version

- DigitalGeorgetown Home
- Georgetown University Institutional Repository
- Georgetown University Medical Center
- Biomedical Graduate Education
- Department of Biostatistics, Bioinformatics & Biomathematics
Graduate Theses and Dissertations - Biostatistics, Bioinformatics & Biomathematics

Creators Titles By Creation Date
Search within this collection:
Browse All Items
Most Recent Submissions

Deconvolution Analysis and Differential Expression Inference for Bulk Tissues and Spatial Transcriptomics

Contributions to Parametric and Semiparametric Structural Equation Modeling Methods with Applications in the Biomedical Sciences

Enhanced Doubly Robust Estimation for Group Comparisons in Presence of Missing Data and Time-varying Confounders

Functional Data Analysis and its Application in Biomedical Research

Effective Semiparametric Analysis for Causal Inference with Time-to-Event Outcomes

- Bodleian Libraries
- Oxford LibGuides
- Bioinformatics
- Theses & Dissertations
Bioinformatics: Theses & Dissertations
- Journals and Conference Proceedings
- Online resources
Links for Theses and Dissertations
- Proquest Dissertations and Theses Search US theses and dissertations. Accessed through OxLip+, search for 'dissertations and theses'.
- Oxford Research Archive (ORA) Search for and download recent Oxford DPhil theses. Also contains an archive of articles, papers and research posters produced by academics and researchers at Oxford University. more... less... ORA is freely available and does not require a log-in.
- EThOS Access to UK theses from the British Library [Currently unavailable]. more... less... To use this service you will be required to set up an individual account.
- DART-Europe Search European E-theses.
Theses and Dissertations On-line
Electronic collections.
A number of recent theses and dissertations prepared at Oxford are available to download from the Oxford Research Archive (ORA) . The British Library provides access to UK theses through its EThOS service . Already digitised UK theses can be downloaded freely as PDF files. Requests can be made to digitise older theses, but there is a cost of around £40 and waiting time of 30 days for digitisation. The British Library no longer provides theses on microfilm.
Finding Oxford Theses
SOLO allows you to search for Theses in the Oxford collections.
1. Navigate to the SOLO homepage.
2. Click on the ' Advanced Search ' button
3. Click the ' Resource Type ' menu and choose the ' Theses ' option.
4. Type in the title or author of the thesis you are looking for and click the ' Search ' button.
Other Relevant Guides
- ORA: Oxford University Research Archive by Jason Partridge Last Updated Apr 10, 2024 2767 views this year
- << Previous: Online resources
- Last Updated: Jun 20, 2024 4:42 PM
- URL: https://libguides.bodleian.ox.ac.uk/bioinformatics
Website feedback
Accessibility Statement - https://visit.bodleian.ox.ac.uk/accessibility
Google Analytics - Bodleian Libraries use Google Analytics cookies on this web site. Google Analytics anonymously tracks individual visitor behaviour on this web site so that we can see how LibGuides is being used. We only use this information for monitoring and improving our websites and content for the benefit of our users (you). You can opt out of Google Analytics cookies completely (from all websites) by visiting https://tools.google.com/dlpage/gaoptout
© Bodleian Libraries 2021. Licensed under a Creative Commons Attribution 4.0 International Licence
Your browser is unsupported
We recommend using the latest version of IE11, Edge, Chrome, Firefox or Safari.
Richard and Loan Hill Department of Biomedical Engineering
Colleges of engineering and medicine, ms in bioinformatics.
Required Semester Hours: 36
Thesis track Heading link Copy link

The thesis track is designed for MS in Bioinformatics students who are interested in conducting research. This track is strongly advised if you may be interested in pursuing a PhD in the future.
Researching and writing a master’s thesis is an academically intensive process that takes the place of 8 credits of traditional coursework. Students work with a faculty advisor to choose a topic of interest, engage in high-level study of that topic, and develop a paper that is suitable for presentation at a conference or submission to a journal.
The thesis experience provides definition to your master’s degree experience and can bolster your application for jobs or doctoral-level study by demonstrating your capabilities.
In the thesis option, you will earn 8 credits in BME 598 Master’s Thesis Research and at least 28 credit hours from coursework. At least 12 of your coursework credits must come from courses at the 500 level, excluding BME 595, BME 596, and BIOE 598. You may be allowed limited credit hours from BME 596 Independent Study with department approval. There is no comprehensive examination.
Recent UIC master’s thesis projects in bioinformatics include:
thesis titles Heading link Copy link
Nikita Dsouza
Strategies for Identification of Small Molecule Inhibitors of Ad2 E3-19K/HLA-A2 Binding Interaction
A Statistical Framework for GeneSet Enrichment Analysis based on DNA Methylation and Gene Expression
Navya Josyula
Identifying Ligand Binding Sites of Proteins using Crystallographic Bfactors and Relative Pocket Sizes
Non-thesis track Heading link Copy link
In the non-thesis track, you earn all of your required 36 credit hours from coursework. Of these, 16 must be from courses at the 500 level. There is no comprehensive examination.
Across-the-board requirements Heading link Copy link
- 1 hour of BME 595
- Present at least one seminar (BME 595) before graduation
- Students entering the program without an undergraduate degree in bioengineering or biomechanical engineering must also take BME 480, BME 481, and BME 530
MS alumni in their own words Heading link Copy link

Daiqing Chen ’21 MS in Bioinformatics
What led you to choose bioinformatics for your MS degree? How do you think computational technology is changing biomedical engineering? I was doing molecular biology during my undergrad. Wet lab experiments are very time- and money-consuming. I have seen people using bioinformatics methods to solve biological questions, and I want to be able to use them. I actually don’t know much about engineering, but I believe a computational method can be useful for any field. The high efficiency allows people to do more things than ever before.
What are your plans for once you have completed your degree? I am planning on working as a research assistant in biological lab, most likely doing research about cancer. My time at UIC helped me get more familiar with American culture.
Have you worked in any labs? Yes, the Computational Functional Genomics Laboratory . I did a project to validate machine learning models that predict kidney function decline. I also worked on high-throughput single-cell sequence analysis.
Your primary hobby/outside interest: Playing badminton.
Favorite restaurant in Chicago: Minhin’s cuisine for the dim sum.
Additional information Heading link Copy link
- MS in Bioinformatics course checklist: thesis track
- MS in Bioinformatics course checklist: non-thesis track
- MS in Bioinformatics graduate catalog page
- UIC Graduate College admissions
- Important deadlines for BME graduate students
- Director’s Welcome
- Participating Departments
- Frontiers in Computational Biosciences Seminar Series
- Current Ph.D. Students
- Current M.S. Students
- Bioinformatics Department Handbook
- B.I.G. Summer Institute
- The Collaboratory
- Diversity and Inclusiveness
- Helpful Information for Current Students
- Joint UCLA-USC Meeting
- Student Blog and Twitter Feed
- Social Gatherings
- Introduction to the Program
- Bioinformatics Admissions Information
- Admissions FAQs
- Student Funding
- Curriculum and Graduate Courses
- Research Rotations
- Qualifying Exams
- Doctoral Dissertation
- Student Publications
- Admissions Information
- Capstone Project
- Undergraduate Courses
- Undergraduate and Masters Research
- Bioinformatics Minor Course Requirements
- Bioinformatics Minor FAQs
- Bioinformatics Minor End-of-Year Celebration
- For Engineering Students
Every master’s degree thesis plan requires the completion of an approved thesis that demonstrates the student’s ability to perform original, independent research.
Students must choose a permanent faculty adviser and submit a thesis proposal by the end of the third quarter of study. The proposal must be approved by the permanent adviser who served as the thesis adviser. The thesis is evaluated by a three-person committee that is nominated by the program and appointed by the Division of Graduate Education. Students must present the thesis in a public seminar.
Featured News
Researchers awarded $4.7 million to study genomic variation in stem cell production, dr. nandita garud recognized for her research on gut microbiome, ucla study reveals how immune cells can be trained to fight infections, ucla scientists decode the ‘language’ of immune cells, dr. eran halperin elected as fellow of international society for computational biology, upcoming events, labor day holiday, recent student publications.
RECENT STUDENT PUBLICATIONS LINK-PLEASE CLICK!
Updates Coming Soon!
Thesis or Dissertation
Each graduate student in the program will work on a dissertation project under dual mentorship, consisting of a primary advisor who is Program Training Faculty, and a co-advisor who may or may not be Program Training Faculty, but must be from a different disciplinary area.
It is expected that the student will meet at least annually with the committee to update the members on his or her progress. As a partial fulfillment for the PhD degree, the student will submit a complete dissertation to be evaluated by a doctoral committee chosen by his or her mentors in consultation with the bioinformatics steering committee. The doctoral dissertation will be submitted to each member of the doctoral committee at least four weeks before the final examination. The student will defend his or her final thesis after the committee's evaluation and will pass or fail depending on the committee's decision.
- PLEASE READ: FAQ on scheduling exams
- UCSD Writing Hub's services for graduate students, including the Dissertation Writer's Workshop
Aarhus University logo
Bioinformatics Research Centre
Master's thesis in bioinformatics.
In the Master’s program in bioinformatics, you must do a 30 ECTS Master’s thesis. You must start your 30 ECTS thesis no later than February 1 (or September 1 ) a year and a half after commencement of your studies (i.e. February 2021 for students admitted in summer 2019, or September 2021 for students admitted in winter 2020). You must complete your thesis (including the exam) no later than June 30 the same year, if you started on February 1 (or January 31 the following year, if you started on September 1).
You can read the course description for the MSc thesis project at:
kursuskatalog.au.dk/en/course/114372/Thesis-30-ECTS-Bioinformatics
You can read some general information and advice about Master’s thesis work at:
https://studerende.au.dk/en/studies/subject-portals/bioinformatics/masters-thesis/masters-thesis/
You can see abstracts of (some) Master's theses from BiRC at:
https://www.birc.au.dk/~cstorm/birc-msc/birc-msc.html
Thesis contract
Before you start your thesis, you must make a thesis contract. The thesis contract must be completed and approved by January 15 (or August 15 ). You can read about how to submit the contract on the above www page. As part of the thesis contract, you must attach a pdf file containing project description, project goals, activity plan, and supervision plan. This is very much like what you have to describe for a Project in Bioinformatics. At BiRC, you should use the following template for this description.
Problem statement, activity plan, and supervision plan (in docx format)
When formulating the thesis project, you should keep in mind that it should cover 30 ECTS of work, i.e. full-time work for the entire semester and the following exam period. Group projects should of course cover this for every group member.
Choosing a topic
Before you can make a thesis contract, and commence your thesis work, you must (of course) chose a topic and a supervisor. The supervisor must be a tenured researcher associated to BiRC, but you can also have one or more co-supervisors.
When choosing a thesis topic, it is a good idea to think about the classes and projects that you have done during your Master’s studies, and what kind of work do you like? Contact potential supervisors as early as possible to discuss your wishes and ideas. Remember that you are always welcome to come by our offices and discuss. You can also ask potential supervisors for examples of thesis’s that they have supervised in order to get a better idea of how a thesis can look.
Also, we plan an information meeting for students that focus on thesis and project work every Fall. Below are the slides from the last such information meeting.
Slides from MSc info meeting (November 2023)
Ten simple rules for writing a great MSc thesis at BiRC (November 2022)
The slides also contain good advice about how to organize your thesis work. The above www page also contains some advice.
Group projects: It is possible to do the thesis project as a group project. Each group member must fill out individual contracts stating the other groups members. A group hand in a single thesis, but each group member is examined individually. In general, we very much encourage group assignments as it for many students is motivating to work together in a group, and to have group member to discuss and solve the many the details of a thesis project together with.
Projects involving external collaborators: It is possible to do a project that involves external collaboration, e.g. with people from industry, or from other university departments. Such collaborators will be associated to your thesis as co-supervisors. In the thesis contract, it is possible to indicate that the thesis project is done in collaboration with an industrial partner, if an NDA has been signed, and if the final thesis report must be made public available.
The thesis report presents the completed work and can be written in Danish or English. The report must contain an English summary/abstract. The summary/abstract is included in the assessment, and the assessment places emphasis on the academic content, as well as the student’s spelling and writing skills. The extent of the thesis report is agreed with the supervisor, but is typically about 50-60 pages excluding frontpage, table of content and appendices. If the MSc thesis is done as a group project provided, the report must be done in such a way that the group members can be assessed individually. This means that you can either (1) do a joint report in which everyone is equally responsible for all parts of the report, or (2) do a joint report, where it is stated (fx in the table of content) who of you has done the individual parts of the report and is responsible for them. See https://studerende.au.dk/en/studies/subject-portals/bioinformatics/masters-thesis/masters-thesis/ under "Group assignment" for details.
In your thesis contract, you state the hand in date. This can between June 1 and 15 (or January 1 and 15 ), earlier dates are also possible. The exact date is (of course) decided in collaboration with your supervisor. You hand in your thesis via Digital Exam (like you are used to for Projects in Bioinformatics).
The thesis exam is 60 min oral exam. It starts with a 30 min presentation from you about your thesis work followed by a 30 min discussion between you, the examiner (your supervisor), and an external examiner. Your presentation is based upon a question that you get from your supervisor one week before the exam. The exam must be held before June 30 (or January 31 ). In principle, the exam can be held from the day after you hand in your thesis. The exact date is decided upon by your supervisor, and often depends on the availability of external examiners. The final grade reflects an overall assessment of your report, your presentation, and your discussion.
If you have any questions about thesis work, then you are always welcome to ask!

Master of Science in Bioinformatics and Computational Biology (BICB-MS)
Starting Fall 2024 the Master’s degree is changing to Bioinformatics Data Science (BIDS-MS)
Program of Study forms:
- MS in Bioinformatics & Computational Biology – Life Sciences (Spring 2024 and earlier)
- MS in Bioinformatics & Computational Biology – Computational Sciences (Spring 2024 and earlier)
- PSM in Bioinformatics & Computational Biology – Life Sciences (Spring 2024 and earlier)
- PSM in Bioinformatics & Computational Biology – Computational Sciences (Spring 2024 and earlier)
The MS degree prepares students for advanced research. The Computational Sciences Concentration allows students with strong quantitative sciences backgrounds to gain knowledge and research experience in developing computational methods and bioinformatics tools and databases for the study of biological systems. The BICB-MS graduates will have solid knowledge and research experience to pursue further study towards a PhD or other professional degree such as MD, MBA or law, or a research career in academia, industry, or government agencies.
The Master’s degree requires 31 credits of course work and must include 15 credits of Bioinformatics Data Science core courses, 3 credits of Ethics, 6 credits of Elective and 1 credit of Seminar.
| (31 Credit Hours Total) | |
|---|---|
| Bioinformatics & Computational Biology Core | 15 Credits |
| Ethics Core | 3 Credits |
| Electives | 6 Credits |
| Seminar (3 semesters) | 1 Credit |
Up to six credits of MS Thesis must be used to meet the degree requirements. The University requirements for master’s thesis shall apply to the thesis in this degree and shall be supervised by the Thesis Faculty Advisor.
| Thesis (6) | |
|---|---|
| Thesis (6) | BINF 869 Master’s Thesis (1-6) |
Up to six credits of Special Problems can be used to meet the degree requirements. The Special Problems credits must be related to the program objectives in bioinformatics data science, and approved by the Graduate Program Committee.
| Non-Thesis (6) | |
|---|---|
| Non-Thesis [Select Two] | Additional Electives (1-6) |
| BINF 666 Special Problems (1-6) |
Bioinformatics Data Science Core Courses
Students must take 15 credits of breadth courses, one course in each of the following areas:
| Bioinformatics and Computational Biology Core (15 Credit Hours) | |
|---|---|
| Bioinformatics | BINF644 Bioinformatics (3) |
| Systems Biology [Select One] | BINF694 Systems Biology I (3) |
| BINF695 Computational Systems Biology (3) | |
| Database [select one] | BINF640 Databases for Bioinformatics (3) |
| CISC637 Database Systems (3) | |
| Biostatistics [select one] | STAT656 Biostatistics (3) |
| STAT611 Regression Analysis (3) | |
| Intro to Discipline [select one] | Computational Sciences Concentration |
| BISC609 Molecular Biology of the Cell (3) | |
| BISC654 Biochemical Genetics (3) | |
| PLSC636 Plant Genes and Genomes (3) | |
| Life Science Concentration | |
| BINF690 Programming for Bioinformatics (3) | |
Elective Courses
Students must take two courses (6 credits). Students are encouraged to explore graduate courses (600-level or higher) in other areas such as electrical engineering, mathematics, linguistics, statistics, and business and economics.
See Elective courses
Ethics Courses
Students must take one of the following ethics core courses (3 credits)
- PHIL655 The Ethics in Data Science and AI
- BUAD840 Ethical Issues in Global Business Environments
- PHIL648 Environmental Ethics
- UAPP650 Values Ethics and Leadership
Students must take three semesters of seminar (two 0 credit; one 1 credit) and give a presentation in their final semester.
| Seminars (1 Credit Hour) | |
|---|---|
| Seminar | BINF 865 Seminar (0-1) |
Master’s Thesis
Up to 6 credits of Master’s Thesis (BINF869) must be used to meet the degree requirements. The master’s thesis shall be supervised by the Thesis Faculty Advisor. Students will prepare and present a research proposal to their Thesis Committee for review and approval of the proposed research project. Following completion of the research outlined in the proposal, the MS degree candidates will prepare a written thesis according to the guidelines set forth by the Graduate College . A thesis defense, preceded by a seminar, will be held. The student’s Faculty Advisor and Thesis Committee will administer and evaluate the thesis defense.
Thesis Committee Guidelines
Formation of Thesis Committee – Students must assemble their thesis committee one month prior to their second academic year. The committee must contain at least three members, with at least two CBCB Affiliate Faculty . Students must complete the Thesis Committee Formation form and complete a one page document proposing a topic and a plan of work. Upon completion, both documents need to be submitted to the Associate Director.
Thesis Committee Meetings – The thesis committee will meet prior to the first semester of the second academic year to discuss project guidelines and assess student progress. If needed, the committee can chose to meet again (prior to the thesis defense) to ensure student is meeting expectations. Within two weeks of the committee meeting, the Thesis Committee Evaluation form must be completed and submitted to the Education and Outreach Coordinator.
Thesis Defense – The thesis defense will be scheduled for the second semester in the second year. Students should note the thesis submission deadlines provided by Office of Graduate and Professional Education and ensure enough time is allotted between the submission deadline and the thesis defense to make appropriate changes and obtain signature pages. The University Thesis and Dissertation Manual must be followed.
The student must submit their thesis to the thesis committee two weeks before the thesis defense date. Within one week of the thesis defense, the thesis committee must submit the Results of the MS Defense form to [email protected]. The original will remain in the student’s file.
Department of Mathematics and Computer Science
Service navigation.
- Privacy Policy
- Accessibility Statement
- DE: Deutsch
- EN: English
- Studying Bioinformatics
Path Navigation
- Bioinformatics
- Master’s Degree Program
- Master's Thesis
Master's Thesis
Master’s thesis with accompanying colloquium (30 credits).
The master’s thesis is meant to prove the student’s ability to work independently on an advanced problem from the bioinformatical field using scientific methods, as well as the student's ability to evaluate the findings appropriately and to depict them both orally and in written form in an adequate manner. (SPO 2019, § 9)
If the study regulations of 2012 apply to you, please have a look here .
If you're looking for a thesis , here are some suggestions.
Unofficial Extract from the Regulations:
- Students can only be admitted to the master's thesis if they have successfully completed modules totaling 60 credits or more within the master's degree program.
- For the registration of the master thesis please use the form "Registration for the master thesis". You can find it on the pages of the examination office ! Important: Be sure to register your master thesis right at the beginning of work! Otherwise you risk that the examiner combination or the topic will not be accepted!
- The master's thesis should be approximately 70 pages in length.
- The processing time is 23 weeks . Note: An extension is not possible. If your thesis is delayed for an important reason (for which you are not responsible), please contact the Examination Office with the relevant supporting documents.
- The written part must be written in English.
- The master's thesis must be evaluated by two authorized examiners . One of the two examiners should be the supervisor of the master's thesis. At least one of the two authorized examiners* must be involved in teaching the master's program and simultaneously be a lecturer at the Department of Mathematics and Computer Science or the Department of Biology, Chemistry, Pharmacy of the Freie Universität Berlin or at Charité.
- If approved by the examining board, the work on the master's thesis can also be done externally at a suitable business or scientific or research institution, as long as scientific and scholarly supervision by an examiner in the program in bioinformatics is ensured.
- The master's thesis is accompanied by a colloquium , which usually takes place in the assigned working group during the processing time. Students are expected to give a one-time presentation lasting approximately 30 minutes on the progress of their master's thesis.
- The master's thesis must be submitted in electronic form (PDF), by e-mail to the examination office. When submitting the thesis, the student must certify in writing that he or she has written the thesis independently and has not used any sources or aids other than those specified. Use the Declaration of Authorship provided by the examination office for this purpose.
*These are usually all PhD scientists involved in teaching in the Master's program in Bioinformatics. However, persons who are not directly involved in teaching may also be authorized. In case of doubt, please contact the examination office , which can check if a certain examiner or combination of examiners is possible or not. Note: The two examiners of a master thesis should come from different working groups.
The Informationen & Anleitungen of the examination office offer further information concerning the registration and submitting regulations of the master’s thesis (in german). The registration form is available in English.
Please note: If you have completed all the coursework and only need to finish the master's thesis, you no longer need to be enrolled, (but you are allowed to, of course).
Every summer semester the Mentoring organizes the workshop “How to write a bachelor’s / master’s thesis in bioinformatics”. Here you receive helpful tips and are free to ask your questions.
Here you can find a compilation of important information (FAQ Abschlussarbeit, in German).
Related Links

Direct Links
JLU von A-Z
Informationen für
- Schülerinnen & Schüler
- Studieninteressierte
- Studierende
- Menschen mit Fluchthintergrund
- Unternehmen
- Jobs & Karriere
- Wissenschaftler/innen
- Promovierende
- Weiterbildungsangebote für JLU-Angehörige
- Lehrerfortbildung
- Wissenschaftliche Weiterbildung
- Ehemalige (Alumni)
- E-Campus ( Stud.IP , ILIAS , FlexNow , eVV )
Studium & Campus
- Vor dem Studium
- Studienangebot
- Bewerbung/Einschreibung
- Information/Beratung
- Vorlesungsverzeichnis
- Studien- und Prüfungsordnungen (MUG)
- Hochschulrechenzentrum
- Universitätsbibliothek
- Campusplan | Geschosspläne/JLUmaps
- Raumvergabe (ZLIS)
- Studierendenwerk/Mensen
- Corporate Design, Leitfäden, Logos
- Bildergalerie Pressestelle
- Formulare | Rundschreiben
- SAP & JustOS (JLU-Online-Shop)
- Rechtliche Grundlagen (MUG)
- Störungsmeldung
- Datenschutz
Karriere, Kultur, Sport, Marketing
- Allgemeiner Hochschulsport (ahs)
- Botanischer Garten
- Career Services
- Gender & JLU
- Hochschuldidaktik
- Justus' Kinderuni
- Sammlungen der JLU
- Universitätsorchester
- Uni-Shop/Merchandising
- E-Mail-Kontakt
- Telefonbuch
- Wegbeschreibung
- Call Justus
Open thesis topics
Within our group we can offer various topics in the field of applied bioinformatics, high-throughput data analysis, genome and metagenome research as well as postgenomics and systems biology. Below you can find a list of suggested open topics for BSc and MSc theses and student projects. For further details on each topic or alternative projects please contact us.

Comparative genome analysis of Streptococcus agalactiae (GBS) from elephants (M.Sc.)
Background Group B Streptococci are fairly common. In livestock, they are the causative agent of an udder inflamation, most often seen in dairy cows.
In elephants, S. agalactiae is associated with Paronchya. Under human care, elephants are known to reach a high age. This comes with an age-related decline in their immune system, which can lead usually harmless skin- or foot diseases to become chronic. Gaining a better knowledge about the bacterial infections is a vital foundation for optimized treatments and therapeutic approaches.
In a newer study done by the "Hessische Landeslabor" (Hesse state labratory (LHL)), some S. agalactiae isolates were compared, using microbiological methods and had extensive biochemical profiles created. Noticable was the high number of isolates, for which the serotypes could not be determined. For this reason some isolates got sequenced, so a full comparative genome analysis could be done, using the latest methods in bioinformatics.
Thesis aims
- Implementation of typical bioinformatic analyses (Assembly, mapping, annotation...)
- Comparative analysis of GBS Isolates (ABR, pan- and coregenome, virulence factors...)
- Closer inspection of Genes for serotyping
Prerequisites
- Interested in solving biological/veterenary questions by usage of bioinformatics
- Extensive knowledge of the Linux command line
- Ability to work independently and methodical
Contact: Linda Fenske
Workflow Design (Nextflow) (M.Sc.)
Analysing (bacterial) sequence data for biological/medical questions means often repeating certain standard processes (QC, Assembly, Annotation etc.)
For better reproduceability and simplification of these processes, flexible pipelines with a wide palette of tools are used. Often Nextflow (of similar workflow tools) is used to enable support for a variety of enviroments or to simplify the installation.
With DSL2, Nextflow recently introduced a significant development of the Nextflow language, which promises a better scalability and modulariziation of pipelines, along with a better design of workflows.
- Revision and updating of an existing workflow for analysing bacerial data
- Transmission of the workflow from nf-DSL1 to DSL2
- Visualising the results (creating a GUI)
Prerequisites
- Knowledge of Nextflow or motivation to become acquainted with Nextflow
- Programming knowledge in Python, Groovy (Nextflow) or similar
- Knowledge and interest in visualisation and processing of data
Platon Bioinformatics Tool Enhancement for Faster Plasmid Identification (M.Sc.) - taken
Modern high-throughput sequencing devices enable the rapid determination of sequence data obtained from interacting microbial communities without a prior cultivation step. Hereby, access to genetic information from otherwise unculturable microbiota is easily achieved. (Computational) Interpretation of such data relies on either assignment of raw sequencing reads to corresponding source organisms in order to infer their taxonomic origin or gene-coding content, or, these metagenome datasets can be assembled, thereby recovering longer contiguous DNA stretches of the underlying microbial genomes.
Assembled metagenomic contigs are typically clustered (most often, depending on coverage or nucleotide composition), yielding individual draft or complete genomes of novel bacterial species. In this process, however, contigs of non-chromosomal origin such as plasmids are often overlooked.
Still, the analysis of plasmids is of utmost imoprtance, since they constitute a key mechanism of horizontal gene transfer between microbial hosts. They are known to harbor essential genes that are beneficial or important for microbial fittness or survival under certain environmental conditions (e.g. in the presence of certain antimicrobial agents) or perform metabolic processes that they otherwise wouldn‘t have been able to (e.g. degradation of novel substrates).
Several bioinformatics applications have been developed for the computational identification of plasmid-borne contigs, most typically focusing on the extraction of plasmid contigs from the assemblies of individual draft genomes. Among these tools are Platon (Schwengers et al., 2020), PlasClass (Pellow et al., 2020) and PlasFlow (Krawczyk et al., 2018), of which Platon exhibits excellent performance, but its runtime characteristics currently impede its application to potentially large metagenome assemblies.
- Overhaul of the Platon code base, switching from a contig-centered approach to one based on bulk data processing in order to significantly decrease overall runtime.
- Inlining of certain sub-analysis steps such as circularity testing into the python codebase instead of relying on the invocation of external tools: (Pyrodigal, pyHMMER, PyTrimal)
- Conditional tool execution: Do not invoke additional tools if preceding steps already exclude a sequence from being a plasmid
- Runtime and performance assessment with regard to the original implementation
Requirements
- Familiarity with Linux and (modular) python programming (incl. unit testing)
- Methodological way of working
- Able to work independently
Contact: Oliver Schwengers
Develop and Compare Curare Modules for Different DGE Libraries (M. Sc)
Differential gene expression analysis (DGE) is a commonly used method in RNA sequencing, in which the expressions of different genes in samples from different conditions are statistically compared to identify relevant genes in stress or defense situations. To simplify the execution of these analyses, the software Curare was developed.
Currently, the R library DESeq2 is used for the statistical evaluation of expression data, but there are also alternative libraries such as edgeR or Limma that pursue similar or completely different statistical approaches.
This Master's thesis aims to write, compare, and combine Curare modules for various DGE libraries. This requires working with different R libraries, integrating the evaluation into Curare (written in Snakemake), and visualizing the results in an HTML report.
- Write Curare modules for different DGE libraries and compare and combine them.
- Learn about different R libraries for statistical analysis of expression data.
- Integrate the analysis in Curare (written in Snakemake) and visualize the results in an HTML report.
Contact: Patrick Blumenkamp
Reconstruction and visualization of KEGG metabolic pathways in the EDGAR platform (M.Sc.)
EDGAR is a web-based platform for analyzing microbial data. It is developed by employees of the Bioinformatics and Systems Biology department at JLU Giessen and provides multifaceted methods for investigating genomes.
KEGG ( Kyoto Encyclopedia of Genes and Genomes) provides curated databases and resources for (among other things) the functional annotation and classification of genes. In previous projects, KEGG functional categories for all organisms and their corresponding genes were computed in the EDGAR platform. These are currently displayed directly in two analysis modules, in purely quantitative terms.
MinPath is a program for reconstructing biological/metabolic pathways. It attempts to infer a minimal biological metabolic network by excluding redundant metabolic pathways that can explain the genes found in a given dataset. The above-mentioned KEGG categories will be used as input for this program.
The goal of the project is to develop a comparative analysis module, based on KEGG pathway information, for the EDGAR platform.
Thesis Aims
- Parse the available KEGG data in a structured manner and compute KEGG metabolic pathways for all given genomes in EDGAR using MinPath.
- Design comparative visualizations for the EDGAR frontend using the resulting data, allowing users to interactively explore their data (see fig. 4 here as an example)
- Adjust the project scope in consultation with the student depending on the project status to accommodate shared ideas, as EDGAR incorporates a wide selection of data with potential for creative analysis methods.
Requirements
Programming skills in Python and JavaScript (can also be learned during the process)
Basic SQL database knowledge
PlasmidHunter: Validation of a metagenome-based plasmid search using public plasmid sequences (M.Sc.)
Plasmids play an important role in the genetic variability of organisms. They replicate independently and between organisms - within and between species. Therefore, plasmids are key drivers of horizontal gene transfer. Often, they are the effective and only difference between commensal and pathogenic bacterial strains. In recent years, it became obvious that plasmids belong to the main mechanisms for the dissemination of antimicrobial resistances and hence are of special interest in medical microbiology. Detecting plasmids and analyzing their dissemination is an important epidemiological and scientific topic that might help to detect current and prevent future outbreaks of antibiotic resistances.
One promising data source containing known and unknown plasmids are whole-metagenome datasets of samples from different sources (soil, waste water, the human gut). For many of these samples, sequencing data is freely accessible in public databases, often annotated with additional meta information such as date, source and location of each sample.
Our project processes these datasets from the MGnify database in a standardized way via modern cloud technologies and makes them accessible to users for a fast search of new plasmids within this huge amount of data.
This master thesis should validate this search via existing plasmid databases (such as PLSDB) and analyze search results including comprehensive visualizations.
- Implementation of a workflow to process PLSDB entries with our existing search workflow
- Statistical analysis of the results, and screen for potential interesting candidates for further analysis
- Visualization of the results
- Knowledge of command line tools and Python
- Interest in cloud technologies
- Prior experience with workflow systems, like Nextflow or Snakemake
Contact: Sebastian Beyvers
Webservice for searching gene families in plants (M. Sc.)
The input is a list of protein sequences. In step 1a, a Pfam search is performed with the sequences to find common domains. In step 1b, a multiple sequence alignment of the sequences is calculated. The conserved regions are automatically extracted from the alignment to calculate HMMs. In step 2, the HMMs of the domains from 1a and 1b are used to search a database of plant proteins.
- The results are visualized and made available for download
- Steps 1 and 2 are also provided as a command-line tool
- The programming language(s) and frameworks can be freely chosen
- Test data will be provided
Contact: Oliver Rupp
R ibosomal binding site prediction based on 16S-rRNA (M.Sc.)
Bacterial translation is initiated by the assembly of ribosomal proteins as part of the translation initiation complex at the coding sequence (CDS) start site. For most CDS, there is a ribosomal binding site (RBS) immediately upstream of the gene, consisting of a 5-10bp spacer and a (partial or complete) Shine-Dalgarno sequence (SD) 5’-AGGAGG-3’ to which the ribosome binds. However, some genes have neither an SD nor a known RBS and are still expressed (Omotajo, D. et al. , 2015) . The Shine-Dalgarno sequence was first described in E. coli but is found in many bacterial genomes and is complementary to the anti-SD sequence at the 3′-end of 16S-rRNA.
The exact Shine-Dalgarno and spacer sequences vary between bacterial species. However, because the anti-Shine-Dalgarno sequence is present in the 16S-rRNA of each bacterial genome, it can be used to predict RBS in a species-independent manner. Therefore, a deep learning approach using the 16S-rRNA sequences and the sequence upstream of the CDS is promising for accurately predicting the presence of RBS independent of species-specific variants.
- Design and implementation of a neural network for ribosomal binding site prediction in bacteria,
- evaluation of the features used by the neural network, and
- analysis of the presence of RBS in exemplary bacterial genomes
- Prior experience with deep learning frameworks such as Tensorflow/Keras, or willingness to learn them
- Prior experience in the development of documented code and dependency management or willingness to learn them
Contact: Julian Hahnfeld
Integrative Omics FAIR Workflow (M.Sc.) Background
Processing and analysing 'omics data often requires applying predefined building blocks of code, i.e. for performing quality control, statistical analysis or machine learning. However, biologists and ecologists are often overwhelmed with the technical complexity of programmatic approaches and interfaces. Hence, scientific workflows can not just automate, but also facilitate important re-occuring processes in high-throughput 'omics analysis.
The existing modularized iESTIMATE pipeline aims at automating and facilitating the complex analysis of ecological metabolomics data and the integration with other phenomics and preparation for sequencing and (meta-)genomics data. The central aim of the pipeline is to extract so called molecular traits that explain molecular mechanisms in plants or microorganisms. Thesis Aims
- Revision and modularisation of existing code to create the R package "iESTIMATE"
- Implementing a workflow in NextFlow or Common Workflow Language (CWL) using test data, implementing unit tests and capture provenance information
- Publish R package and the workflow following the FAIR principles
- Knowledge of R and a bit of Python
- Knowledge of Linux command line, containers, NextFlow (Groovy), YAML, or motivation to become acquainted with them
- Keen interest in analysis of integrative 'omics data and in topics in molecular ecology
Contact: Kristian Peters
Bioinformatics, Master of Science
Zanvyl krieger school of arts and sciences, ms in bioinformatics, joint offering with the whiting school of engineering.
Johns Hopkins University offers an innovative graduate program that prepares professionals for success in bioinformatics. Drawing from the strengths of the Krieger School of Arts and Sciences and the Whiting School of Engineering, this program fully integrates the computer science, bioscience, and bioinformatics skills and knowledge needed to pursue a career in this dynamic field.
The 11-course degree program is thesis-optional and can be completed part-time or full-time and onsite, online, or through a combination of onsite and online courses.
Admissions Criteria for All Advanced Academic Programs
Program-specific requirements.
In addition to the materials and credentials required for all programs, the Master of Science in Bioinformatics requires an undergraduate degree in the biological sciences or engineering with at least a 3.0 on a 4.0 scale.
- Statement of purpose: Please provide a statement, up to one page in length, describing your personal background and/or a part of your life experience that has shaped you or your goals. Feel free to elaborate on personal challenges and opportunities that have influenced your decision to pursue a graduate degree at Johns Hopkins.
- Two semesters of organic chemistry
- One semester of biochemistry
- One semester of an introduction to programming using Java, C++, C, or Python
- One semester of data structures
- One semester of probability/statistics
- One semester of calculus
Program Requirements
Students in the MS in Bioinformatics program must complete 11 courses:
- Two required core courses
- Seven customizable core courses
- One elective from bioscience
- One elective from computer science
After completing the above courses, students may choose an independent study project (optional).
| Code | Title | Credits |
|---|---|---|
| Core Courses - Required: | 8 | |
| Molecular Biology | ||
| Epigenetics, Gene Organization & Expression | ||
| Core Courses - Customizable | 11 | |
| Introduction to Bioinformatics | ||
| Biological Databases and Database Tools | ||
| Practical Computer Concepts for Bioinformatics | ||
| Principles of Database Systems | ||
| Algorithms for Bioinformatics | ||
| Foundations of Algorithms | ||
| Select four of the following: | 16 | |
| Bioinformatics: Tools for Genome Analysis | ||
| Protein Bioinformatics | ||
| Molecular Phylogenetic Techniques | ||
| Next Generation DNA Sequencing and Analysis | ||
| Gene Expression Data Analysis and Visualization | ||
| Advanced Practical Computer Concepts for Bioinformatics | ||
| Advanced Genomics and Genetics Analyses | ||
| Practical Introduction to Metagenomics | ||
| Genomic and Personalized Medicine | ||
| Linked Data and the Semantic Web | ||
| Neural Networks | ||
| Principles of Bioinformatics | ||
| Computational Genomics | ||
| Computational Drug Discovery,Dev | ||
| Statistics for Bioinformatics | ||
| Modeling and Simulation of Complex Systems | ||
| Algorithms for Structural Bioinformatics | ||
| Systems Biology | ||
| Applied Machine Learning | ||
| Electives | ||
| Computer Science | ||
| Select one of the following: | 3 | |
| Foundations of Software Engineering | ||
| XML Design Paradigms | ||
| Principles and Methods in Machine Learning | ||
| Data Visualization | ||
| Principles of Enterprise Web Development | ||
| Mobile Application Development for the Android Platform | ||
| Software Systems Engineering | ||
| Large-Scale Database Systems | ||
| Advanced Machine Learning | ||
| Evolutionary and Swarm Intelligence | ||
| Independent Project in Bioinformatics | ||
| Big Data Processing Using Hadoop | ||
| Biotechnology | ||
| Select one of the following: | 4 | |
| Advanced Cell Biology | ||
| Cellular Signal Transduction | ||
| Human Molecular Genetics | ||
| Principles of Immunology | ||
| Virology | ||
| Molecular Basis of Pharmacology | ||
| Genes & Disease | ||
| Gene Therapy | ||
| Emerging Infectious Diseases | ||
| Cancer Biology | ||
| Clinical & Molecular Diagnostics | ||
| Clinical Trial Design and Conduct | ||
| Recombinant DNA Laboratory | ||
| High Throughput Screening & Automation Lab | ||
| Independent Research in Biotechnology | ||
| Total Credits | 42 | |
You may select other electives with the approval of your adviser
See course listings page for the Center for Biotechnology Education
See course listings page for Computer Science
MS in Bioinformatics with Thesis Option
Students interested in pursuing the MS in Bioinformatics with the thesis are required to take 12 courses. The thesis requires a two-semester research project. Students complete AS.410.800 Independent Research in Biotechnology first and AS.410.801 Biotechnology Thesis the following semester. Students interested in this option should consult with the program director or their academic adviser.
Learning Outcomes
Students in this program will:
- Critique current and classic research in molecular biology
- Search public databases in order to analyze data in a biological context
- Implement sequence alignment tools to elucidate the deeper context of biological data
- Develop bioinformatics tools to address biological problems
- Write computer programs to build databases within a biological context in multiple computer languages
- Design deployable computer algorithms
- Develop skills to meet individual career goals in computational biology and related fields.

Tips & Tricks
Current research topics in bioinformatics.
Researchers working in the scientific area always want to explore new and hot topics to make informed choices. In this article, all new, current, and demanding research topics in bioinformatics are mentioned. This article is helpful for the researchers who are looking for trends in bioinformatics to select a research topic of broad-spectrum.
Since the research in bioinformatics and its applications
are exponentially increasing every year, it is essential to know hot topics for researchers who are trying to make a career in this area. Currently, most of the research is focused on treating deadly diseases such as “ cancer, coronary artery disease, HIV, chronic infections ”, and so on . In silico drug designing is always demanding in designing inhibitors or potential drugs for such diseases. Besides, a lot of scientists are working on next-generation sequencing, big data , and cancer . A recent study has found that the interest of researchers in these topics plateaued over after the early 2000s [1].
Besides the above mentioned hot topics, the following topics are considered demanding in bioinformatics.
- Cloud computing, big data, Hadoop
- Machine learning
- Artificial intelligence
- Functional genomics
- Rna-seq analysis (equally relevant along with next-generation sequencing techniques)
- Data mining (including text search, data integration, database development, and management)
- Neural networks
- Mathematical modeling
- Mirna function identification
- Evolutionary studies
- Genomics, transcriptomics, and proteomics
- Metabolomics
If you are new and trying to learn bioinformatics, then read the following articles:
- Bioinformatics- Where & How to Start?
List of Bioinformatics Books for Beginners
- Hahn A., Mohanty S.D., Manda P. (2017) What’s Hot and What’s Not? – Exploring Trends in Bioinformatics Literature Using Topic Modeling and Keyword Analysis. In: Cai Z., Daescu O., Li M. (eds) Bioinformatics Research and Applications. ISBRA 2017. Lecture Notes in Computer Science, vol 10330. Springer, Cham. https://doi.org/10.1007/978-3-319-59575-7_25
Careers in Bioinformatics and Computational Biology
Md simulation using gromacs: things to remember.
The Team at Bioinformatics Review includes top notch bioinformaticians and scientists from across the world. Visit our Team page to know more.

You may like
You must be logged in to post a comment Login
You must be logged in to post a comment.
CMake installation and upgrade: What worked & what didn’t?!
CMake is a widely used cross-platform build system that automates the process of compiling and linking software projects. In bioinformatics, CMake can be utilized to manage the build process of software tools and pipelines used for data analysis, algorithm implementation, and other computational tasks. However, managing the versions of CMake or upgrading it on Ubuntu (Linux) can be a trivial task for beginners. In this article, we provide methods for installing and upgrading CMake on Ubuntu.
(more…)
Common mistakes made during computational docking.

Computational docking is not a trivial task once we avoid making some mistakes. In this section, let’s learn about some important points that we should keep in mind while performing computational docking.
How to download FASTA sequences from PDB for multiple structures?

In this article, we are going to download FASTA sequences for multiple structures from PDB [1]. We need to have PDB IDs only for input. (more…)
How to install the LigAlign plugin on Pymol on Ubuntu (Linux)?

Few errors appear when we try to run the LigAlign plugin [1] in Pymol [2]. For example, if you try to run the ligand_alignment plugin, it will give you multiple errors including “ Unable to initialize LigAlign v1.00 “, or “ can’t run LigAlign v1.00 ” or “ incorrect Python syntax ” or “ Plugin has been installed but initialization failed “. In this article, we explain the reason for this issue and how you can rectify these errors. (more…)
How to make an impactful science presentation?
After your hard work, it is time to showcase your study and the methods of your study to an audience. You must make every point useful and informative. Here, in this article, we are going to share some tips to make your scientific presentation impactful. (more…)
Basic bioinformatics concepts to learn for beginners
This article is for beginners who are stepping into the field of bioinformatics. We will discuss some basic concepts that you need to learn while trying to enter the field of bioinformatics. (more…)
A Beginner’s Guide on How to Write Good Manuscripts

Drafting a manuscript could be a difficult task for beginners in the field of research. In this article, we will provide a few tips for how can you write a good manuscript being a research scholar. (more…)
How to remove HETATMS and chains from PDB file?
This is a basic tutorial on removing the hetero-atoms (HETATMS) and chains from PDB files. It is an essential step for computational and molecular dynamics simulation. (more…)
Importance of Reasoning in Research

Research is considered complicated. As it involves reading multiple research articles and reviews, devising hypotheses and appropriate experiments, and last but not the least, getting a significant output. This seems difficult to read multiple research papers and then extract useful information regarding your project. It involves a methodology to read and understand research articles in one go. This is explained in our previous article “A guide on how to read the research articles” . This article explains the importance of thinking during your entire research. (more…)
How to Find Binding Pocket/ Binding Site for Docking?
Finding binding sites/pockets in a target protein is one of the important steps in docking studies. It is relatively easier to find a binding pocket in the proteins whose resolved structures are available in PDB than that of the predicted structures. In this article, we will discuss the ways to know a binding pocket or a binding site in a target protein. (more…)
Bioinformatics is an interesting field of research combining biological sciences and computer sciences. In this article, we will discuss making careers in bioinformatics for starters. (more…)
MD Simulation
Molecular dynamics (MD) simulation is considered amongst the important methods in bioinformatics. Installation of MD simulation software and execution of their commands is critical. It requires several parameters to be considered before performing simulations. A single mistake may result in impractical outputs. In this article, we will discuss such important things to remember during the MD simulation and installation and execution of its software (GROMACS) [1,2]. (more…)
Bioinformatics Books
It is difficult to decide which book to read to start learning bioinformatics. Beginners can read this article to know the basic steps involved in learning bioinformatics. A few books are suggested in this article to read for starters in bioinformatics. (more…)
Bioinformatics- Where & How to Start?
Bioinformatics being an interdisciplinary area of biological science and computer science may sound complicated to beginners in this field. However, it is quite simple. The only thing you need is knowledge in both areas. Here is a way for the beginners to start with bioinformatics. (more…)
Common mistakes made during Autodock Vina Installation and Execution

Several errors occur while installing MGLTools and Autodock Vina on Ubuntu. We have explained the complete process of Autodock Vina installation and docking in previous articles. Here are some common errors and mistakes that should be taken care of while installing and running Vina on Ubuntu. (more…)
What does a bioinformatician do?
Bioinformaticians play an important part in data analysis and result interpretation in the field of bioinformatics. However, it is unclear to many what specific role bioinformaticians play day to day. We are often asked by many about what exactly a bioinformatician does. But first, who is a bioinformatician?
MGL Tools & Autodock Vina installation: Frequently Asked Questions and Answers
I have received several e-mails from researchers and students alike regarding installing MGL Tools and Autodock Vina on Ubuntu. Most questions are similar in nature, so I thought of answering them once and for all. In this article, I have collected some frequently asked questions and provided the link to their answers in our question-answer section of Bioinformatics Review. (more…)
A guide on how to read the research articles
Reading a research article could be a problem for students starting their careers in research. Research papers are sometimes difficult to understand especially when you are new to this field. In this article, we will discuss how to read and understand a research article. (more…)
Bioinformatics and programming languages- what do you need to know!
There are various things which come to mind when someone is going to enter in the field of Bioinformatics and the topmost concern is “Do I need to learn computer languages to pursue my career in Bioinformatics?”. The answer is a bit tricky but it could be both “yes” and “no”. This article will describe the conditions where you need to learn programming languages in Bioinformatics. (more…)
Site-specific docking: Frequently Asked Questions & answers for starters

I have been getting several E-mails from researchers and students alike regarding in-silico docking. Most questions are similar in nature, so I thought of answering them once and for all. In this article, I have collected some frequently asked questions and provided the link to their answers present in our question-answer section of Bioinformatics Review.
It is good to have questions in mind and they can be solved in a way as quoted by Sir Einstein:
“We cannot solve our problems with the same thinking we used when we created them.”
In this article, I have collected some of the most Frequently Asked Questions while performing site-specific and/ or blind docking. You have to consider a lot of factors before performing an actual docking on a protein with a specific ligand.
Question: How do you predict protein’s binding sites?
Question: What is the difference between the blind docking and binding site based docking?
LATEST ISSUE

Course Catalog
Bioinformatics, ms.
for the degree of Master of Science in Bioinformatics
Students pursing this major must choose one of these concentrations:
Animal Sciences Crop Sciences Computer Science Information Sciences
The MS degree can be taken in a thesis or non-thesis format, depending on the department. For either format, the research adviser must be affiliated with the Bioinformatics program.
Admission Applicants must hold a bachelor's degree equivalent to that granted by the University of Illinois Urbana-Champaign. The recommended background for graduate students entering the Bioinformatics degree program is a bachelor's or master's degree in life sciences, computer and mathematical sciences, or engineering, with a minimum of five hours of molecular and cell biology, six hours of general chemistry, nineteen hours of mathematics and statistics, and three hours of introduction to computing. Prerequisites vary somewhat for the different departmental concentrations. Students should view the web page of the specific department they wish to apply to for detailed information about admission criteria and degree requirements. Those links are below:
- Department of Animal Sciences
- Department of Computer Science
- Department of Crop Sciences
- School of Information Sciences
Financial Aid Fellowships, research assistantships, and teaching assistantships (all of which include tuition and partial fee waivers) are awarded on a competitive basis by the admitting department. All applicants, regardless of U.S. citizenship, whose native language is not English and who wish to be considered for teaching assistantships must submit minimum test scores as determined by university policy.
Bioinformatics Program Bioinformatics website
Contact the individual departments listed below Animal Sciences Crop Sciences Computer Science Information Sciences
Admissions Graduate College Admissions Requirements
Print Options
Send Page to Printer
Print this page.
Download Page (PDF)
The PDF will include all information unique to this page.
2024-2025 Catalog (PDF)
A copy of the full 2024-2025 catalog.
Get the Reddit app
## A subreddit to discuss the intersection of computers and biology. ------ A subreddit dedicated to bioinformatics, computational genomics and systems biology.
Am I overthinking my Master Thesis?
to give you a bit of context: I am currently studying in a bioinformatics Master and expect to do my Thesis next semester. Over the last year I have been working for a research group in the immunology field mainly applying machine learning models.
Unfortunately I felt like I had hit a ceiling on what they could teach me since the majority, including the group leader in the group have no informatics background whatsoever.
Since its my Master Thesis I didn't want to do some task I had done over the whole last year without learning anything new. I talked to the group leader and she could not really offer me anything concrete. I general I had always the feeling she just saw me as a guy who can quickly code here something together before getting to the more interesting immunology stuff.
So a week ago I told her I won't extend my contract and do my Thesis somewhere else. I am currently talking to a former colleague who offered me a Thesis position at the company he is working. So I am thinking this might be a great opportunity to see how work in the industry compares to academia. But this position seems more centered around software dev and less data science/bioinformatics and I am unsure as of yet which area interests me more.
So long story short: Am I overthinking the importance of the actual Thesis topic? In the end the degree is what counts right? Did anyone have similar situations and how did you decision turn out?
We use cookies to make your experience on this website better.
Accept Cookies
College of Sciences and Mathematics Homepage
- Toggle Search
- Find People
"Analyzing Demographic Preparedness to Disaster Types and Severity: Insights from Human Mobility Data"
Thesis Defense by Dikshya Panta
In the News
- UB Directory
- Giving to UB >
- Find Your Cause >
Coming Soon to a Lab Near You

By Dirk Hoffman
Published October 18, 2023
The Department of Microbiology and Immunology has two new faculty members starting later this year who are eager to recruit new members to their research labs.
Multidisciplinary Approaches to Studying Gut Bacteria
Yolanda Yue Huang, PhD , will be joining the Jacobs School of Medicine and Biomedical Sciences as an assistant professor of microbiology and immunology on Dec. 1.
She comes from Lawrence Berkeley National Laboratory where she worked in the group of Adam Arkin, PhD, as an Astellas Pharma awardee of the Life Sciences Research Foundation postdoctoral fellowship.
The laboratory is a U.S Department of Energy Office of Science national laboratory managed by the University of California. There, she developed a novel high-throughput functional genomic approach to study gut bacteria.
Huang grew up in Canada and completed her Bachelor of Science degree in biochemistry at McGill University.
She then pursued a doctoral degree in chemical biology in the lab of Emily Balskus, PhD, at Harvard University. Her thesis work uncovered a new radical enzyme responsible for anaerobic amino acid metabolism.
“What makes this pathway interesting is that the amino acid is predominantly sourced from the host — diet and abundant host proteins. This highlights how microbes have evolved to metabolize abundant nutrients available in the gut environment,” Huang says.
One challenge in the microbiome field is that most microbes have not been characterized.
“The amount of sequencing data is increasing exponentially, but it is really difficult to translate this data into biological functions. I am excited to tackle this knowledge gap by leveraging multidisciplinary approaches in my group,” Huang says.
Specifically, the Huang lab will combine high-throughput functional genomics, bioinformatics, biochemistry, and microbiology to rapidly connect genes to phenotypes for characterizations at the molecular level.
Another focus of the group will be to study how bacteriophages (bacterial viruses) influence bacterial functions and composition dynamics. Phages encode an even greater proportion of unknown genetic information and their role in the gut is not well understood.
“I am super excited to embark on the next chapter of my career and to be joining the vibrant scientific community at UB. I especially look forward to mentoring trainees at all levels and enabling them in their career paths,” Huang says.
For more information about the Huang lab, contact Huang at [email protected] .
Research Focused on Bacterial Pathogens
Ryan C. Hunter, PhD , is an associate professor of microbiology and immunology, who will be joining the Jacobs School faculty on a full-time basis Nov. 23.
He received his Bachelor of Science degree from the University of Guelph in Canada in 2001. He went on to pursue postbaccalaureate research at NASA’s Jet Propulsion Laboratory prior to earning his doctoral degree in microbiology 2007 under the direction of Terry J. Beveridge, PhD, at the University of Guelph.
His graduate work focused on the microbial adaptation to their growth environments, their role in metal redox transformations, and their broader impacts on global elemental cycling.
Subsequently, Hunter was awarded a Canadian Cystic Fibrosis Foundation postdoctoral fellowship for studies at the Massachusetts Institute of Technology and was named a HHMI postdoctoral scholar at the California Institute of Technology in the lab of Dianne K. Newman, PhD.
Hunter and Newman used a multidisciplinary approach to define the in vivo chemical environment of the cystic fibrosis airways, and how bacterial pathogens adapt to and co-evolve with the host over time.
In 2012, Hunter received a National Institutes of Health Pathway to Independence Award (K99/R00) and joined the faculty in the Microbiology department at the University of Minnesota in 2013.
Since the start of his independent career, Hunter’s research has focused on the in vivo physiology of bacterial pathogens and how they obtain nutrients from the host.
He has a particular interest in mucus-microbe interactions, and manipulating those interactions to shape our microbiota in many disease contexts (cystic fibrosis, chronic sinusitis, periodontal disease, and GI complications including colorectal cancer).
The Hunter lab opens its doors in the Department of Microbiology and Immunology at the Jacobs School in November 2023.
For more information about the Hunter lab, contact Hunter at [email protected] .

IMAGES
VIDEO
COMMENTS
This project will assess whether AMGs generally evolve into distinct shorter versions of the bacterial gene and whether the transfer of metabolic genes from phages to bacteria is a prevalent phenomenon. To this end, publicly available genomes of phages and bacteria will be scanned for metabolic genes (Shaffer et al. 2020).
List of PhD theses produced at the Bioinformatics Laboratory or under co-supervision of the Bioinformatics Laboratory.
MSc thesis: In the Bioinformatics group, we offer a wide range of MSc thesis projects, from applied bioinformatics to computational method development. Here is a list of available MSc thesis projects. Besides the fact that these topics can be pursued for a MSc thesis, they can also be pursued as part of a Research Practice.
Theses. Thesis Preparation and Filing: Staff from the University Archives and the UCLA Graduate Division present information on University regulations governing manuscript preparation and completion of degree requirements. Students should plan to attend at least one quarter before they plan to file a thesis or dissertation. More information is ...
Bioinformatic analysis of next-generation sequencing data Master`s Thesis Bioinformatics Masters Degree Programme, Institute of Biomedical Technology
Thesis Advisors must: Hold a faculty appointment at a Harvard University school at the rank of Assistant Professor or above. Have a research program that uses computational methods in biomedical applications. Students may be co-advised by up to two advisors, with approval from the Program. The Thesis Advisor is expected to meet with students ...
Research in bioinformatics, or interdisciplinary investigation of biomedical problems with significant bioinformatic components. This research is at the master's level, leading to completion of a scientific project for presentation as a thesis. May be repeated for credit.
In this thesis, I have used 16S sequencing data from mock bacterial communities to evaluate the sensitivity and specificity of several bioinformatics pipelines and genomic reference libraries used for microbiome analyses, with a focus on measuring the accuracy of species-level taxonomic assignments of 16S amplicon reads.
Functional Data Analysis and its Application in Biomedical Research . Li, Haiou (Georgetown University, 2023) The objective of the dissertation is to develop new statistical methods for functional data analysis motivated by several biomedical research. In many applications with functional observations, the main goals of statistical ...
A number of recent theses and dissertations prepared at Oxford are available to download from the Oxford Research Archive (ORA). The British Library provides access to UK theses through its EThOS service. Already digitised UK theses can be downloaded freely as PDF files. Requests can be made to digitise older theses, but there is a cost of ...
The thesis track is designed for MS in Bioinformatics students who are interested in conducting research. This track is strongly advised if you may be interested in pursuing a PhD in the future.
The Master of Science in Bioinformatics non-thesis option is a Professional Science Master's degree program. The mission of this professionally oriented program is to train graduates for leadership roles in bioinformatics, biotechnology, biomedicine and other sectors of the life sciences. The program imparts interdisciplinary knowledge ...
My passion to work with bioinformatics and molecular biology had been fulfilled by getting involved in this master thesis project. It was a great opportunity for me to practice bioinformatics techniques and the wet-lab work which enabled me to gain an immense knowledge that is useful for my future research.
Thesis. Every master's degree thesis plan requires the completion of an approved thesis that demonstrates the student's ability to perform original, independent research. Students must choose a permanent faculty adviser and submit a thesis proposal by the end of the third quarter of study. The proposal must be approved by the permanent ...
The doctoral dissertation will be submitted to each member of the doctoral committee at least four weeks before the final examination. The student will defend his or her final thesis after the committee's evaluation and will pass or fail depending on the committee's decision.
Master's Thesis in Bioinformatics. In the Master's program in bioinformatics, you must do a 30 ECTS Master's thesis. You must start your 30 ECTS thesis no later than February 1 (or September 1) a year and a half after commencement of your studies (i.e. February 2021 for students admitted in summer 2019, or September 2021 for students ...
The MS degree prepares students for advanced research. The Computational Sciences Concentration allows students with strong quantitative sciences backgrounds to gain knowledge and research experience in developing computational methods and bioinformatics tools and databases for the study of biological systems. The BICB-MS graduates will have solid knowledge and research experience to pursue ...
The master's thesis is meant to prove the student's ability to work independently on an advanced problem from the bioinformatical field using scientific methods, as well as the student's ability to evaluate the findings appropriately and to depict them both orally and in written form in an adequate manner. (SPO 2019, § 9) Please read § 9 ...
Open thesis topics. Within our group we can offer various topics in the field of applied bioinformatics, high-throughput data analysis, genome and metagenome research as well as postgenomics and systems biology. Below you can find a list of suggested open topics for BSc and MSc theses and student projects.
Zanvyl Krieger School of Arts and Sciences 2024-25 Edition
Researchers working in the scientific area always want to explore new and hot topics to make informed choices. In this article, all new, current, and demanding research topics in bioinformatics are mentioned. This article is helpful for the researchers who are looking for trends in bioinformatics to select a research topic of broad-spectrum. Since the […]
Bioinformatics, MS. for the degree of Master of Science in Bioinformatics. Students pursing this major must choose one of these concentrations: Animal Sciences Crop Sciences Computer Science Information Sciences. The MS degree can be taken in a thesis or non-thesis format, depending on the department. For either format, the research adviser ...
to give you a bit of context: I am currently studying in a bioinformatics Master and expect to do my Thesis next semester. Over the last year I have been working for a research group in the immunology field mainly applying machine learning models.
Students enrolled in the Infectious Diseases Concentration must work on a thesis project related to infectious diseases (drug discovery, medical devices, bioinformatics, the molecular basis of a disease, etc.). Option A (6.0-credit Master's Research Thesis) and Option B (12.0-credit Master's Research Thesis) are available.
Geosciences Thesis Defense - Dikshya Panta: Time: Jul 16, 2024 (09:00 AM) Location: Zoom Details: "Analyzing Demographic Preparedness to Disaster Types and Severity: Insights from Human Mobility Data"
Her thesis work uncovered a new radical enzyme responsible for anaerobic amino acid metabolism. ... Specifically, the Huang lab will combine high-throughput functional genomics, bioinformatics, biochemistry, and microbiology to rapidly connect genes to phenotypes for characterizations at the molecular level.