
Microbe Notes

The Wobble Hypothesis: Definition, Statement, Significance
There are more than one codon for one amino acid . This is called degeneracy of genetic code. To explain the possible cause of degeneracy of codons, in 1966, Francis Crick proposed “the Wobble hypothesis”.
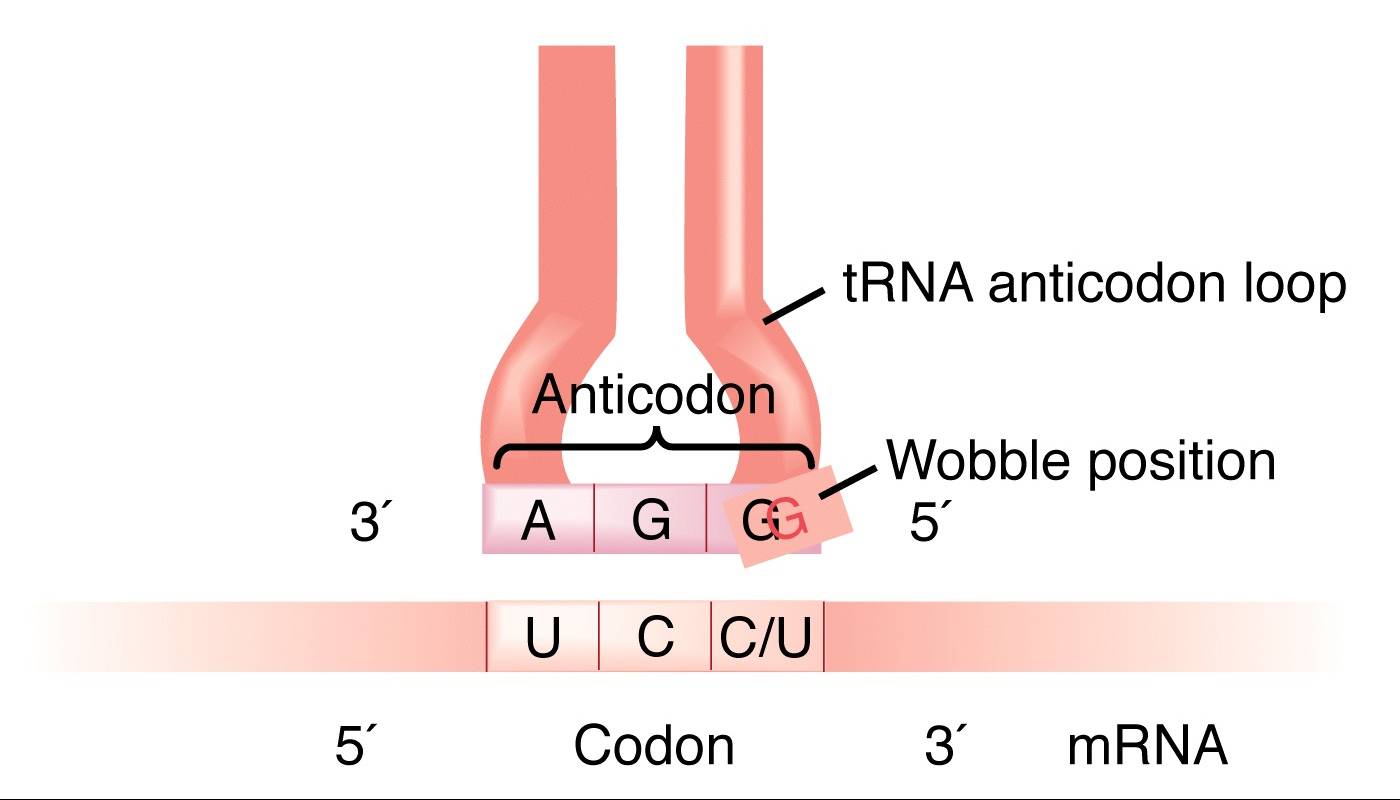
According to The Wobble Hypothesis, only the first two bases of the codon have a precise pairing with the bases of the anticodon of tRNA, while the pairing between the third bases of codon and anticodon may Wobble (wobble means to sway or move unsteadily).
The phenomenon permits a single tRNA to recognize more than one codon. Therefore, although there are 61 codons for amino acids, the number of tRNA is far less (around 40) which is due to wobbling.
Table of Contents
Interesting Science Videos
The Wobble Hypothesis Statement
The wobble hypothesis states that the base at 5′ end of the anticodon is not spatially confined as the other two bases allowing it to form hydrogen bonds with any of several bases located at the 3′ end of a codon.
This leads to the following conclusions:
- The first two bases of the codon make normal (canonical) H-bond pairs with the 2nd and 3rd bases of the anticodon.
- At the remaining position, less stringent rules apply and non-canonical pairing may occur. The wobble hypothesis thus proposes a more flexible set of base-pairing rules at the third position of the codon.
- The relaxed base-pairing requirement, or “wobble,” allows the anticodon of a single form of tRNA to pair with more than one triplet in mRNA.
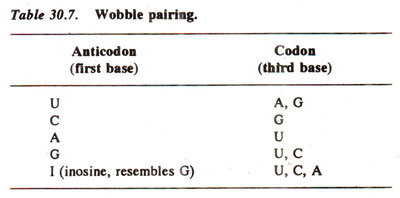
- The rules: first base U can recognize A or G, first base G can recognize U or C, and first base I can recognize U, C or A.
Crick’s hypothesis hence predicts that the initial two ribonucleotides of triplet codes are often more critical than the third member in attracting the correct tRNA.
Wobble base pairs
- A wobble base pair is a pairing between two nucleotides in RNA molecules that does not follow Watson-Crick base pair rules.
- The four main wobble base pairs are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C).
- In order to maintain consistency of nucleic acid nomenclature, “I” is used for hypoxanthine because hypoxanthine is the nucleobase of inosine.
- Inosine displays the true qualities of wobble, in that if that is the first nucleotide in the anticodon then any of three bases in the original codon can be matched with the tRNA.
Significance of the Wobble Hypothesis
- Our bodies have a limited amount of tRNAs, and wobble allows for broad specificity.
- Wobble base pairs have been shown to facilitate many biological functions, most clearly proven in the bacterium Escherichia coli .
- The thermodynamic stability of a wobble base pair is comparable to that of a Watson-Crick base pair.
- Wobble base pairs are fundamental in RNA secondary structure and are critical for the proper translation of the genetic code.
- Wobbling allows faster dissociation of tRNA from mRNA and also protein synthesis.
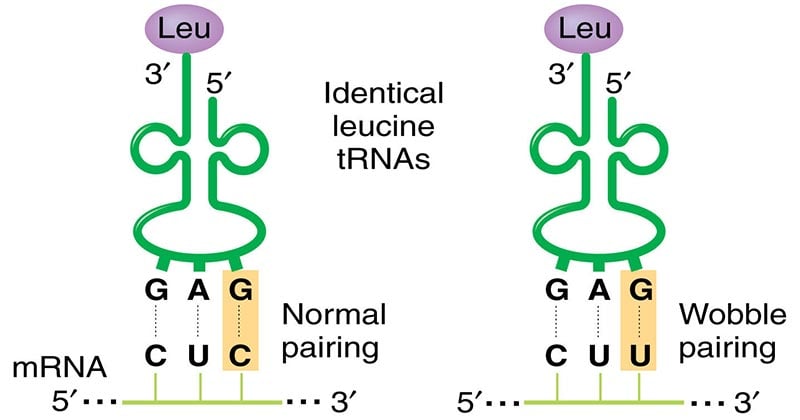
- The existence of wobble minimizes the damage that can be caused by a misreading of the code; for example, if the Leu codon CUU were misread CUC or CUA or CUG during transcription of mRNA, the codon would still be translated as Leu during protein synthesis.
- Verma, P. S., & Agrawal, V. K. (2006). Cell Biology, Genetics, Molecular Biology, Evolution & Ecology (1 ed.). S .Chand and company Ltd.
- Klug, W. S., & Cummings, M. R. (2003). Concepts of genetics. Upper Saddle River, N.J: Prentice Hall.
- https://www.slideshare.net/sweetmerrymindfreak/genetic-code-53764823
- https://www.slideshare.net/ArchaDave/genetic-code-24735057
- https://www.scribd.com/document/344722502/Wobble-hyothesis-pdf
- https://en.wikipedia.org/wiki/Wobble_base_pair#Wobble_hypothesis
About Author
Sagar Aryal
2 thoughts on “The Wobble Hypothesis: Definition, Statement, Significance”
I need help to drow my method how i contact you my email is [email protected]
nice explaination
Leave a Comment Cancel reply
Save my name, email, and website in this browser for the next time I comment.
This site uses Akismet to reduce spam. Learn how your comment data is processed .
The Wobble Hypothesis: Importance and Examples
The “wobble hypothesis” refers to a concept in molecular biology that explains the degeneracy of codons.
Since there are only 20 amino acids and 64 possible codons, multiple codons may code for a single amino acid during protein synthesis. In molecular biology, this redundancy or multiplicity of codons is termed degeneracy.
This hypothesis helps explain how a relatively limited number of tRNA molecules can recognize and bind to multiple codons for the same amino acid, facilitating efficient and accurate protein synthesis. Experimental evidence has supported the wobble hypothesis, a fundamental concept in understanding the genetic code and translation machinery.\
Table of Contents
Importance of Wobble Hypothesis
Overall, the wobble hypothesis plays a fundamental role in understanding protein synthesis and the genetic code, with implications for various aspects of molecular biology, genetics, and biotechnology.
Examples of Wobble Hypothesis
Here are some examples of the wobble hypothesis in action:
Limitation of Wobble Hypothesis
Hello, I am Ashma Shrestha. I had recently completed my Masters degree in Medical Microbiology. Passionate about writing and blogging. Key interest in virology and molecular biology.
We love to get your feedback. Share your queries or comments Cancel reply
This site uses Akismet to reduce spam. Learn how your comment data is processed .
Recent Posts
- BiologyDiscussion.com
- Follow Us On:
- Google Plus
- Publish Now

Wobble Hypothesis (With Diagram) | Genetics
ADVERTISEMENTS:
In this article we will discuss about the concept of wobble hypothesis.
Crick (1966) proposed the ‘wobble hypothesis’ to explain the degeneracy of the genetic code. Except for tryptophan and methionine, more than one codons direct the synthesis of one amino acid. There are 61 codons that synthesise amino acids, therefore, there must be 61 tRNAs each having different anticodons. But the total number of tRNAs is less than 61.
This may be explained that the anticodons of some tRNA read more than one codon. In addition, identity of the third codon seems to be unimportant. For example CGU, CGC, CGA and CGG all code for arginine. It appears that CG specifies arginine and the third letter is not important. Conventionally, the codons are written from 5′ end to 3′ end.
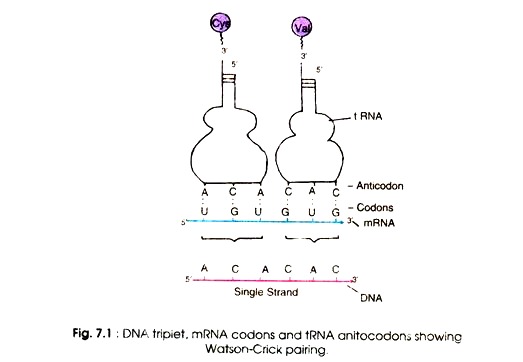
Therefore, the first and second bases specify amino acids in some cases. According to the Wobble hypothesis, only the first and second bases of the triple codon on 5′ → ‘3 mRNA pair with the bases of the anticodon of tRNA i.e A with U, or G with C.
The pairing of the third base varies according to the base at this position, for example G may pair with U. The conventional pairing (A = U, G = C) is known as Watson-Crick pairing (Fig. 7.1) and the second abnormal pairing is called wobble pairing.
This was observed from the discovery that the anticodon of yeast alanine-tRNA contains the nucleoside inosine (a deamination product of adenosine) in the first position (5′ → 3′) that paired with the third base of the codon (5′ → 3′). Inosine was also found at the first position in other tRNAs e.g. isoleucine and serine.
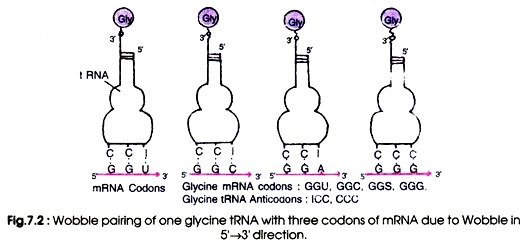
The purine, inosine, is a wobble nucleotide and is similar to guanine which normally pairs with A, U and C. For example a glycine-tRNA with anticodon 5′-ICC-3′ will pair with glycine codons GGU, GGC, GGA and GGG (Fig 7.2). Similarly, a seryl-tRNA with anticodon 5′-IGA-3′ pairs with serine codons UCC, UCU and UCA (5-3′). The U at the wobble position will be able to pair with an adenine or a guanine.
According to Wobble hypothesis, allowed base pairings are given in Table 7.5:
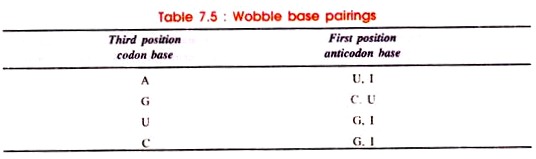
Due to the Wobble base pairing one tRNA becomes able to recognise more than one codons for an individual amino acid. By direct sequence of several tRNA molecules, the wobble hypothesis is confirmed which explains the pattern of redundancy in genetic code in some anticodons (e.g. the anticodons containing U, I and G in the first position in 5’→ 3′ direction)
The seryl-tRNA anticodon (UCG) 5′-GCU-3′ base pairs with two serine codons, 5′-AGC-3′ and 5′-AGU-3′. Generally, Watson-Crick pairing occurs between AGC and GCU. However, in AGU and GCU pairing, hydrogen bonds are formed between G and U. Such abnormal pairing called ‘Wobble pairing’ is given in Table 7.5.
Three types of wobble pairings have been proposed:
(i) U in the wobble position of the tRNA anticodon pairs with A or G of codon,
(ii) G pairs with U or C, and
(iii) 1 pairs with A, U or C.
Related Articles:
- Short Notes on Anticodons | Genetics
- Genetic Code: Degeneracy and Universality | Protein
Microbiology , Genetics , Wobble Hypothesis , Concept of Wobble Hypothesis
- Anybody can ask a question
- Anybody can answer
- The best answers are voted up and rise to the top
Forum Categories
- Animal Kingdom
- Biodiversity
- Biological Classification
- Biology An Introduction 11
- Biology An Introduction
- Biology in Human Welfare 175
- Biomolecules
- Biotechnology 43
- Body Fluids and Circulation
- Breathing and Exchange of Gases
- Cell- Structure and Function
- Chemical Coordination
- Digestion and Absorption
- Diversity in the Living World 125
- Environmental Issues
- Excretory System
- Flowering Plants
- Food Production
- Genetics and Evolution 110
- Human Health and Diseases
- Human Physiology 242
- Human Reproduction
- Immune System
- Living World
- Locomotion and Movement
- Microbes in Human Welfare
- Mineral Nutrition
- Molecualr Basis of Inheritance
- Neural Coordination
- Organisms and Population
- Photosynthesis
- Plant Growth and Development
- Plant Kingdom
- Plant Physiology 261
- Principles and Processes
- Principles of Inheritance and Variation
- Reproduction 245
- Reproduction in Animals
- Reproduction in Flowering Plants
- Reproduction in Organisms
- Reproductive Health
- Respiration
- Structural Organisation in Animals
- Transport in Plants
- Trending 14
Privacy Overview
| Cookie | Duration | Description |
|---|---|---|
| cookielawinfo-checkbox-analytics | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Analytics". |
| cookielawinfo-checkbox-functional | 11 months | The cookie is set by GDPR cookie consent to record the user consent for the cookies in the category "Functional". |
| cookielawinfo-checkbox-necessary | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookies is used to store the user consent for the cookies in the category "Necessary". |
| cookielawinfo-checkbox-others | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Other. |
| cookielawinfo-checkbox-performance | 11 months | This cookie is set by GDPR Cookie Consent plugin. The cookie is used to store the user consent for the cookies in the category "Performance". |
| viewed_cookie_policy | 11 months | The cookie is set by the GDPR Cookie Consent plugin and is used to store whether or not user has consented to the use of cookies. It does not store any personal data. |

- __Crossword/Puzzle
- __Resources
- Difference Between
- Biology MCQ
- _Chemistry for Biologists
What is Wobble Hypothesis?
- The multiple codes for a given amino acid( degeneracy )
- Possible suppression of point mutations in the third base of the codon
- More easily removed deacylated tRNA during protein synthesis.
- Fewer tRNA molecules than expected.
Our website uses cookies to improve your experience. Learn more
Contact form


- G.Engineering
- BBTechniques
Wobble Hypothesis
The Wobble Hypothesis , proposed by Francis Crick in 1966, explains the degeneracy of the genetic code. The hypothesis attributes this degeneracy to imprecise pairing between the third base of the codon and the first base of the anticodon on tRNA. The wobble position has two main properties:
- Loose bindingThe third position of the codon has a looser binding between the cognate tRNA and the codon in the mRNA.
- Non-Watson-Crick base pairingsThe third position of the codon can accommodate a variety of non-Watson-Crick base pairings. For example, G may pair with T or U.
- The wobble hypothesis is important in translating the genetic code because it allows a single tRNA to recognize more than one codon. This means that there are fewer tRNAs (around 40) than codons (61) for amino acids.
Wobble Base Pair
- A wobble base pair is type of non-canonical base pairing that occurs between two nucleotides in RNA molecules, the codon and the anticodon of mRNA and tRNA, respectively that does not follow Watson-Crick base pair rules.
- The four main wobble base pairs are guanine-uracil ( G-U ), hypoxanthine-uracil ( I-U ), hypoxanthine-adenine ( I-A ), and hypoxanthine-cytosine ( I-C )
- Wobble base pairs are fundamental in RNA secondary structure and are critical for the proper translation of the genetic code
- The term “wobble” refers to the flexibility or deviation from the standard Watson-Crick base pairing rules at the third position of the codon, which allows a single tRNA to recognize more than one codon for the same amino acid.
- This phenomenon explains the degeneracy of the genetic code and reduces the number of tRNA molecules required for protein synthesis.
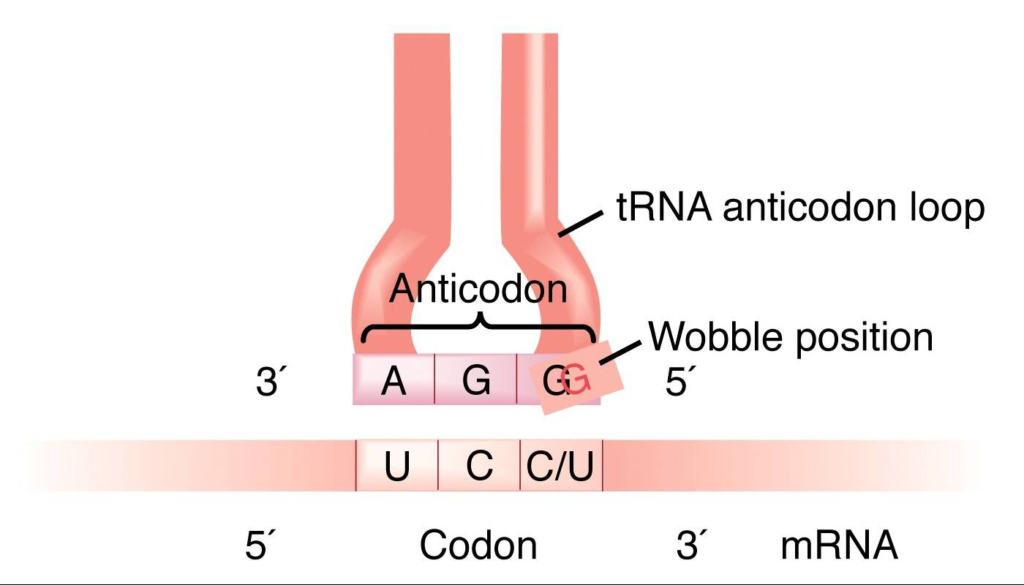
The Wobble Hypothesis
Genetic Code Study Notes:
- There are 64 possible codons in the genetic code, each consisting of a 3-nucleotide sequence. Translation requires tRNA molecules, each with an anticodon that complements a specific mRNA codon. Canonical Watson-Crick base pairing is used for stable tRNA-mRNA binding during translation.
- In the standard genetic code, 3 mRNA codons (UAA, UAG, UGA) act as stop codons, terminating translation. This leaves 61 mRNA codons that require tRNA molecules, suggesting a need for 61 types of tRNA.
- Due to the limited number of tRNA species in organisms (usually fewer than 45), some tRNA types can pair with multiple synonymous codons.
- Francis Crick proposed the Wobble Hypothesis in 1966, suggesting that the 5′ base on the anticodon has non-standard base pairing due to spatial flexibility. The “wobble” at the third codon position allows for small conformational adjustments, influencing the overall pairing geometry of tRNA anticodons.
- Crick suggested that the first two bases of the codon form strong and specific Watson-Crick base pairs with the second and third bases of the anticodon, while the third base of the codon can form weaker and less specific base pairs with the first base of the anticodon.
- The first two bases of each codon are primary determinants of specificity. The third base pairing is not very stable and wobbles. For example, CUU, CUG, CUC, CUA codons, which differ only at the third base represent the same amino acid leucine. The first two bases of the codon form strong base pairs with the corresponding bases of the anticodon but the third base forms weak hydrogen bond.
- This allows some tRNA molecules to bind to more than one codon, as long as they differ only at the third position. For example, a tRNA with the anticodon 5′-GmAA-3′ can recognize both UUC and UUU codons for phenylalanine.
The Wobble Rules
Crick also proposed a set of rules that govern the possible wobble base pairs, based on the geometry and hydrogen bonding patterns of the nucleotides involved. The rules are as follows:
- G can pair with U or C (in addition to the canonical C)
- U can pair with A or G (in addition to the canonical A)
- I (inosine, a deaminated form of A) can pair with A, U, or C
- A can pair only with U (the canonical pair)
- C can pair only with G (the canonical pair)
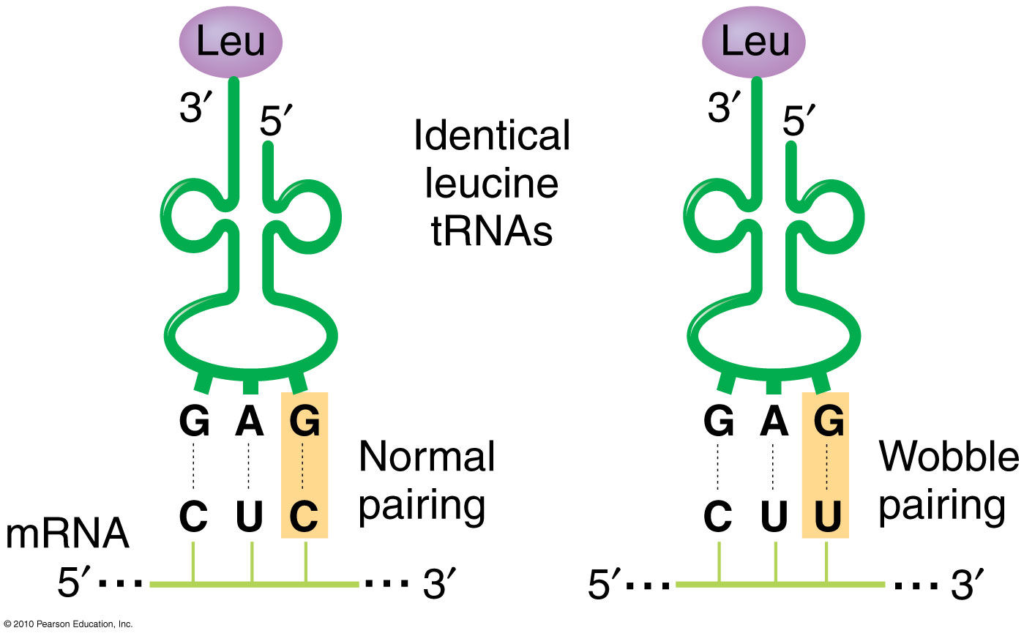
These rules imply that some codons for the same amino acid are more versatile than others in terms of wobble pairing. For instance, codons ending with A or C can be recognized by only one specific tRNA, while codons ending with U or G can be recognized by one or two tRNAs, depending on whether the first base of the anticodon is U, G, or I.
| tRNA 5′ anticodon base | mRNA 3′ codon base (Crick) | mRNA 3′ codon base (Revised) |
|---|---|---|
| A | , C, G, or (A) | |
| C | ||
| G | or U | or U |
| U | or G | , G, U, or (C) |
| I | A, C, or U | A, C, or U |
The Significance of Wobble Base Pairing
Wobble base pairing has several advantages for the cell:
- It reduces the number of tRNA genes and tRNA molecules needed to translate all 61 sense codons, which saves genetic space and energy.
- It allows for some mutations or errors at the third position of the codon without affecting the protein sequence, which increases the genetic robustness and diversity.
- It enables some tRNA molecules to act as suppressors of nonsense mutations by recognizing stop codons and inserting amino acids instead, which may rescue some defective proteins.
Wobble base pairing is a common feature of RNA secondary structure and is essential for the accurate and efficient translation of the genetic code.
Leave a review Cancel reply
Your email address will not be published. Required fields are marked *
Your comment *
Your name *
Your Email *

Sign in to your account
Username or Email Address
Remember Me
Update image
- Wobble hypothesis
Support our developers

More in this section
- The Genetic Code
- Properties of genetic code
- Chain initiation and chain termination codons
- Synonym codons and degeneracy
- Mutations and the genetic code
- New genetic codes in mitochondria and ciliate protozoa
- Suppressor mutations, base substitutions and suppressor tRNAs
- Second genetic code, and second half of the genetic code
- Recoding of the genetic code
© 2012 - 2024 Biocyclopedia
Disclaimer Privacy Policy Feedback
Login to your account
Login / signup with.

Wobble Pair
- Living reference work entry
- Latest version View entry history
- First Online: 25 December 2022
- Cite this living reference work entry

- Henderson James Cleaves 11 , 12 , 13
29 Accesses
A wobble pair, or wobble-base pair, is a hydrogen-bonded pairing between two nucleotides generally occurring between two RNA molecules. These base pairs are geometrically distinct from the canonical Watson–Crick-type base-pairing. The main wobble base pairs are hypoxanthine-uracil (I-U, where I represents inosine, the nucleoside formed from hypoxanthine), guanine-uracil (G-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C). Wobble base pairs are of comparable thermodynamic stability to Watson–Crick base pairs. Wobble base pairs occur frequently in RNA secondary structure and are important for proper translation of the genetic code.
In 1966, Francis Crick proposed the Wobble hypothesis to account for the fact that most organisms do not seem to have as many tRNA molecules as would be required for complete translation of the genetic code. There are 64 possible codons in the genetic code. During translation, each of these codons requires a tRNA molecule with a...
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
Institutional subscriptions

Author information
Authors and affiliations.
Earth-Life Science Institute (ELSI), Tokyo Institute of Technology, Tokyo, Japan
Henderson James Cleaves
Blue Marble Space Institute of Science, Washington, DC, USA
Center for Chemical Evolution, Georgia Institute of Technology, Atlanta, GA, USA
You can also search for this author in PubMed Google Scholar
Editor information
Editors and affiliations.
Laboratoire d'Astrophysique de Bordeaux, University of Bordeaux, Pessac CX, France
Muriel Gargaud
Department of Astronomy, University of Massachusetts, Amherst, MA, USA
William M. Irvine
Centro Biología Molecular Severo Ochoa, Univ. Autónoma de Madrid Cantoblanco, Madrid, Spain
Ricardo Amils
Analytical-, Environm., Geo-Chemistry, Vrije Universiteit Brussel, VUB, Brussels, Belgium
Philippe Claeys
Earth-Life Science Institute, Tokyo Institute of Technology, WASHINGTON, DC, USA
Radio Astronomy, Paris Observatory, Paris, France
Maryvonne Gerin
Observatoire de Paris-Meudon, Meudon, France
Daniel Rouan
International Space Science Institute, Bern, Bern, Switzerland
Tilman Spohn
Centre Francois Viète, Universite de Nantes, Nantes, France
Stéphane Tirard
Innovaxiom, Paris CX, France
Michel Viso
Rights and permissions
Reprints and permissions
Copyright information
© 2022 Springer-Verlag GmbH Germany, part of Springer Nature
About this entry
Cite this entry.
Cleaves, H.J. (2022). Wobble Pair. In: Gargaud, M., et al. Encyclopedia of Astrobiology. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-27833-4_5248-2
Download citation
DOI : https://doi.org/10.1007/978-3-642-27833-4_5248-2
Received : 02 September 2021
Accepted : 23 September 2021
Published : 25 December 2022
Publisher Name : Springer, Berlin, Heidelberg
Print ISBN : 978-3-642-27833-4
Online ISBN : 978-3-642-27833-4
eBook Packages : Springer Reference Physics and Astronomy Reference Module Physical and Materials Science Reference Module Chemistry, Materials and Physics
- Publish with us
Policies and ethics
Chapter history
DOI: https://doi.org/10.1007/978-3-642-27833-4_5248-2
DOI: https://doi.org/10.1007/978-3-642-27833-4_5248-1
- Find a journal
- Track your research
- Subscriber Services
- For Authors
- Publications
- Archaeology
- Art & Architecture
- Bilingual dictionaries
- Classical studies
- Encyclopedias
- English Dictionaries and Thesauri
- Language reference
- Linguistics
- Media studies
- Medicine and health
- Names studies
- Performing arts
- Science and technology
- Social sciences
- Society and culture
- Overview Pages
- Subject Reference
- English Dictionaries
- Bilingual Dictionaries
Recently viewed (0)
- Save Search
- Share This Facebook LinkedIn Twitter
Related Content
Related overviews.
transfer RNA
genetic code
translation
See all related overviews in Oxford Reference »
More Like This
Show all results sharing these subjects:
- Life Sciences
wobble hypothesis
Quick reference.
A theory to explain the partial degeneracy of the genetic code due to the fact that some t-RNA molecules can recognize more than one codon. The theory proposes that the first two bases in the codon and anticodon will form complementary pairs in the normal antiparallel fashion. However, a degree of steric freedom or ‘wobble’ is allowed in the base-pairing at the third position. Thus, for serine, six m-RNA codons may be paired with only three t-RNA anticodons.
From: wobble hypothesis in A Dictionary of Zoology »
Subjects: Science and technology — Life Sciences
Related content in Oxford Reference
Reference entries.
View all reference entries »
View all related items in Oxford Reference »
Search for: 'wobble hypothesis' in Oxford Reference »
- Oxford University Press
PRINTED FROM OXFORD REFERENCE (www.oxfordreference.com). (c) Copyright Oxford University Press, 2023. All Rights Reserved. Under the terms of the licence agreement, an individual user may print out a PDF of a single entry from a reference work in OR for personal use (for details see Privacy Policy and Legal Notice ).
date: 15 July 2024
- Cookie Policy
- Privacy Policy
- Legal Notice
- Accessibility
- [185.39.149.46]
- 185.39.149.46
Character limit 500 /500
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- v.1(1); 2000 Jul 17

The G·U wobble base pair
Gabriele varani.
1 Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK and
William H. McClain
2 Department of Bacteriology, 1550 Linden Drive, E.B. Fred Hall, University of Wisconsin, Madison, WI 53706-1567, USA
The G·U wobble base pair is a fundamental unit of RNA secondary structure that is present in nearly every class of RNA from organisms of all three phylogenetic domains. It has comparable thermodynamic stability to Watson–Crick base pairs and is nearly isomorphic to them. Therefore, it often substitutes for G·C or A·U base pairs. The G·U wobble base pair also has unique chemical, structural, dynamic and ligand-binding properties, which can only be partially mimicked by Watson–Crick base pairs or other mispairs. These features mark sites containing G·U pairs for recognition by proteins and other RNAs and allow the wobble pair to play essential functional roles in a remarkably wide range of biological processes.
Historical perspective
Little did Francis Crick realize 30 years ago that his G·U wobble base pair used in decoding mRNA codons would have such an expansive biological significance ( Crick, 1966 ). When considering codon–anticodon interactions, Crick noticed that the G and U bases would be able to form two hydrogen bonds by interacting through the same face of the base involved in Watson–Crick pairing (Figure (Figure1). 1 ). This prediction was confirmed when the G·U wobble pair in yeast tRNA Phe was observed at atomic resolution ( Ladner et al. , 1975 ; Quigley and Rich, 1975 ). The history of the G·U pair is, in fact, even older. Yeast tRNA Ala , which was the first RNA molecule whose primary sequence was determined, could be folded into a secondary structure containing a single G·U wobble pair ( Holley et al. , 1965 ). It was later shown that the G·U pair is a major determinant of that molecule’s amino acid acceptor identity ( Hou and Schimmel, 1988 ; McClain and Foss, 1988 ). G·U pairs have now been found in virtually every class of functional RNA, and have been shown to play many essential roles that are based upon the unique chemical and structural properties of the wobble pair. Structural features of G·U wobble pairs and other non-Watson–Crick pairs have recently been reviewed elsewhere ( Hermann and Westhof, 1999 ; Masquida and Westhof, 2000 ). Here, we will review the biological roles of G·U pairs and discuss the specific features that allow them to function in a remarkably wide variety of biological contexts.

Fig. 1. Watson–Crick G·C and A·U base pairs differ from wobble G·U pairs in the type and location of functional groups that are projected into major and minor grooves. These pairs also differ in the orientation of the bases with respect to the phosphodiester backbone. Whereas the glycosidic angle is similar (~54°) for all nucleosides in Watson–Crick pairs, both angles for G and U differ in the wobble pair.
Ubiquitous building blocks of RNA structure
The functional importance of G·U pairs is underscored by their frequently high evolutionary conservation. For example, the wobble pair at the third position of the acceptor helix of tRNA Ala is conserved in nearly all living organisms ( Sprinzl et al. , 1996 ). This conservation implies that the G·U pair possesses unique features that cannot be duplicated by any other nucleotide pair. G·U pairs constitute the most common mismatch in the helices of rRNA ( Gautheret et al. , 1995 ), and other tRNAs contain as many as four, but typically one, G·U pairs ( Sprinzl et al. , 1996 ). The mRNAs coding for ribosomal proteins S15 ( Bénard et al. , 1998 ) and L30 ( Li et al. , 1996 ) contain G·U pairs that provide recognition signals for autoregulation of protein synthesis. With the discovery of RNA catalysis, G·U pairs were also found to be associated with this ancient biological function of RNA. In fact, the first class of ribozymes discovered, group I self-splicing introns, contain a G·U pair at the site of cleavage that is nearly universally conserved ( Doudna et al. , 1989 ; Hur and Waring, 1995 ). Replacement of this wobble pair with a Watson–Crick pair compromises reactivity and fidelity ( Doudna et al. , 1989 ; Pyle et al. , 1994 ). The hepatitis delta virus ribozyme also identifies its self-cleavage site through recognition of a G·U wobble pair ( Perrotta and Been, 1996 ; Ferre-D’Amare et al. , 1998 ). Group II self-splicing introns contain a nearly universally conserved G·U pair within domain 5, surrounded by two base pairs that are also nearly invariant ( Peebles et al. , 1995 ; Abramovitz et al. , 1996 ; Konforti et al. , 1998 ). Finally, RNAs that have been selected in vitro from large (10 14 ) pools of randomized RNA sequences to perform various biological functions often contain G·U pairs ( Ciesolka and Yarus, 1996 ; Illangasekare and Yarus, 1999 ; Khvorova et al. , 1999 ).
The G·U pair functions in so many different contexts because it specifies unique recognition sites for proteins as diverse as aminoacyl-tRNA synthetases and ribosomal proteins, for other RNAs and for divalent metal ions. This ability resides in the distinctive structural, chemical and thermodynamic properties of the G·U pair, and in the unique conformational properties it confers upon RNA double helices. Each of these features can be exploited during recognition by proteins and other ligands.
Distinctive chemical, thermodynamic and structural properties
G·U pairs present a unique array of hydrogen bond donors and acceptors in the RNA major and minor grooves. These chemical groups can be recognized by complementary functionalities in a protein, RNA or other ligands. The exocyclic amino group in the minor groove is a distinctive feature of the G·U pair, since it is not base paired and is shifted with respect to the equivalent group in a normal G·C pair (Figure (Figure1). 1 ). Distinguishing chemical features are distributed in the major grooves well, and the presence of co-planar guanosine N7, guanosine O6 and uridine O4 defines a region of deep negative electrostatic potential ( Allain and Varani, 1995a ; McDowell and Turner, 1996 ). In contrast, G·C and A·U pairs both project an NH 2 group into the major groove that interrupts the electrostatic field (Figure (Figure2). 2 ). This steep electrostatic gradient is a major determinant of the ability of divalent metal ions to bind the major groove of G·U pairs, but not that of double helical tracts composed entirely of Watson–Crick pairs ( Ott et al. , 1993 ; Allain and Varani, 1995a ; Konforti et al. , 1998 ). The affinity for metal ions becomes even stronger when two G·U pairs are present side-by-side ( Cate and Doudna, 1996 ; McDowell and Turner, 1996 ; Kieft and Tinoco, 1997 ). Since most RNA enzymes are metalloenzymes in the sense that metal-binding sites generated by unique RNA structures position metals for promoting catalysis ( Pyle, 1993 ), the ability of G·U pairs to bind divalent metal ions is likely to be important for RNA catalysis ( Konforti et al. , 1998 ).

Fig. 2. The G·C, A·U and G·U pairs have different functional groups in the major groove, leading to a significantly more electronegative environment at and near the G·U pair. As shown in this image, the surface electrostatic potential (negative is red and positive is blue) of tRNA Ala acceptor end minihelices differs between the wild-type molecule containing G·U (right) and a mutant molecule where G·U is replaced by G·C (left).
A second important aspect of G·U pair function is its thermodynamic stability, which approaches that of Watson–Crick pairs and exceeds that of most other mispairs ( Jaeger et al. , 1989 ; Mathews et al. , 1999 ; Strazewski et al. , 1999 ). This high thermodynamic stability allows G·U pairs to substitute functionally for Watson–Crick base pairs in phylogenetically conserved double-helical tracts in rRNA, ribozymes and other RNAs. Also, in some sequence contexts, G·U pairs stabilize backbone turns. In tRNA, for example, the G·U pair is most frequently found at the junction of the single-stranded V loop and the T helix where the polynucleotide chain makes a sharp turn ( Clark and Klug, 1975 ). However, thermodynamic stability is not sufficient for G·U function. For example, the aminoacylation capacity of active and inactive mutants of tRNA Ala with various substitutions of G·U does not correlate with their respective thermodynamic stability ( Strazewski et al. , 1999 ).
G·U wobble pairs embedded within A-form RNA helices have a distinctive structure that results from the displacement of the bases of the G·U pair relative to the bases of Watson–Crick pairs. The glycosidic bond angle between the base and C1′ sugar atom of Watson–Crick pairs in an A-form RNA helix is ~54° for each of the four nucleotides (Figure (Figure1). 1 ). Thus, the four standard base pairs can be interchanged at a particular site in a double-stranded region without substantially affecting the helical parameters. In contrast, the glycosidic bond angles are dissimilar in the G·U wobble pair (G, 40° and U, 65°, approximately). The substitution of G·U by any Watson–Crick pair or by a U·G pair therefore results in a structural perturbation in any sequence context in which the wobble pair is located. In addition to these sequence-independent properties, other features of the structure of double helical regions containing wobble pairs depend strongly on sequence context ( Masquida and Westhof, 2000 ). As noted both in crystal ( Westhof et al. , 1985 ; Masquida et al. , 1999 ; Mueller et al. , 1999 ) and in solution structures ( Allain and Varani, 1995b ; Ramos and Varani, 1997 ), G·U pairs introduce a pattern of overtwisting/undertwisting of the RNA double helix. This helical twist is influenced by the identity of the base pairs immediately adjacent to the wobble pair. The conformation of the phosphodiester backbone around the wobble pair also varies between G·U-containing sequences and even between crystallographic environments of a given sequence.
A final distinctive property of G·U base pairs refers to their conformational flexibility. G·U pairs are conformationally soft in the sense that they are found in different conformations in different chemical and structural environments. For example, G·U pairs respond more sharply to sequence context and crystal packing interactions than do Watson–Crick base pairs ( Westhof et al. , 1985 ). Because of this property, the conformation of the RNA double helix can be more easily altered at sites containing wobble pairs, allowing for recognition by induced fit ( Ramos and Varani, 1997 ; Chang et al. , 1999 ). As an example, a significant helical kink is observed at a G·U pair in the aspartyl-tRNA synthetase-bound form of tRNA Asp relative to free, unbound tRNA Asp ( Ruff et al. , 1991 ).
Specific recognition: chemical identity and structural context
As discussed in the previous section, unique chemical features distinguish G·U pairs from Watson–Crick pairs or other mismatches. The functional role of individual chemical groups of the G·U pair in acceptor minihelix tRNA Ala was demonstrated in an in vitro analysis of the enzymic activity of alanyl-tRNA synthetases ( Musier-Forsyth et al. , 1991 ; Musier-Forsyth and Schimmel, 1992 ). These studies identified specific chemical groups in the G·U pair that are critical for alanyl-tRNA synthetase activity. These include the exocyclic amino group of two guanosines, including that of G·U, and several ribose 2′-OH groups, all located in the minor groove of the tRNA acceptor end (Figure (Figure3). 3 ). A related but not identical ensemble of chemical groups was also demonstrated to be important for the ability of the catalytic core of group I introns to recognize the substrate for splicing or to stabilize the enzyme’s transition state ( Strobel and Cech, 1995 ).

Fig. 3. View into the minor groove of RNA double helices showing the exocyclic amino groups (blue) and 2′-OH groups (red) that are critical for the activity of the G·U pairs in different functional contexts. The figure shows the three-dimensional structure of the tRNA Ala acceptor end minihelix (left) and of the P1 helix substrate of group I self-splicing introns (right), highlighting chemical groups in the minor groove that contribute to G·U activity. A set of related but not identical chemical groups at and around the wobble pair is essential for activity in both contexts.
In addition to presenting distinctive chemical groups in the major and minor groove, individual G·U pairs also possess unique conformational features dictated by the sequence context in which the wobble pair is found and by factors that bind to it. The importance of sequence context and of the structural and dynamic features of the wobble pair in its function was demonstrated in genetic and biochemical studies of tRNA Ala function conducted in vivo ( Gabriel et al. , 1996 ; Chang et al. , 1999 ; McClain et al. , 1999 ). These studies identified C·C and C·A nucleotide substitutions that functionally replace the G·U in tRNA Ala even though they contain different chemical features. These results and those from other extensive investigations led to the conclusion that the G·U pair in tRNA Ala is recognized through a combination of the direct read-out of its distinctive chemical identity and by the indirect recognition of the structural features of the double helix in which the G·U pair is embedded. G·U pairs in other functional contexts may also be recognized through a similarly complex mechanism. For example, the guanosine unpaired amino group is energetically important in the function of the group I self-splicing intron ( Strobel and Cech, 1996 ), and other mispairs do not function as well as G·U within the upstream splice site for cleavage. However, A·C, which may be a close structural analogue of G·U, provides the best substitute ( Doudna et al. , 1989 ).
Since the conformation of an RNA double helix at and around the wobble pair varies from one sequence to another, incorporation of a G·U pair and the subsequent sequence-dependent distortion of the RNA double helix leads to an expansion of the structural diversity of A-form RNA. This property allows G·U pairs to form the main anchor points or fulcrums in networks of three-dimensional contacts with the catalytic core of an RNA enzyme or the RNA recognition domain of a protein. These networks of contacts centered on the wobble pair contribute to the high specificity observed in recognition or catalysis. Indirect recognition of G·U is a particularly powerful discrimination mechanism. It allows for productive interaction with cognate ligands, and thus positive discrimination. It also allows for rejection of non-cognate substrates through non-productive interactions with those substrates that lack a required G·U. In addition, a non-cognate substrate can be rejected as a result of steric conflict resulting from the distinctive structure of a G·U pair. An example of the latter type of discrimination is the non-productive interaction between charged selenocysteine tRNA and elongation factor Tu, which results from a particular G·U pair in that specialized tRNA ( Rudinger et al. , 1996 ).
G·U pairs are used with remarkable frequency in biology to expand the chemical and conformational universe accessible to double-stranded RNA and thereby provide unique recognition sites. Protein and RNA enzymes and RNA-binding proteins bind to unique G·U sites by recognizing chemical features common to all G·U pairs and that differ from Watson–Crick and other mismatched pairs. Conformational features of G·U-containing RNA double helices unique to the specific sequence context of each G·U pair provide diversity and allow a variety of unique interaction sites to be built from this otherwise simple recognition tag.
- Abramovitz D.L., Friedman, R.A. and Pyle, A.M. (1996) Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science , 271 , 1410–1413. [ PubMed ] [ Google Scholar ]
- Allain F.H.-T. and Varani, G. (1995a) Divalent metal ion binding to a conserved wobble pair defining the upstream site of cleavage of group I self-splicing introns. Nucleic Acids Res. , 23 , 341–350. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Allain F.H.-T. and Varani, G. (1995b) Structure of the P1 helix from group I self splicing introns. J. Mol. Biol. , 250 , 333–353. [ PubMed ] [ Google Scholar ]
- Bénard L., Mathy, N., Grunberg-Magnago, M., Ehresmann, B., Ehresmann, C. and Portier, C. (1998) Identification in a pseudoknot of a U·G motif essential for the regulation of the expression of ribosomal protein S15. Proc. Natl Acad. Sci. USA , 95 , 2564–2567. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Cate J.H. and Doudna, J.A. (1996) Metal-binding sites in the major groove of a large ribozyme domain. Structure , 4 , 1221–1229. [ PubMed ] [ Google Scholar ]
- Chang K.Y., Varani, G., Bhattacharya, S., Choi, H. and McClain, W.H. (1999) Correlation of deformability at a tRNA recognition site and aminoacylation specificity. Proc. Natl Acad. Sci. USA , 96 , 11764–11769. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ciesolka J. and Yarus, M. (1996) Small RNA-divalent domains. RNA , 2 , 785–793. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Clark B.F.C. and Klug, A. (1975) Structure and function of tRNA with special reference to the three dimensional structure of yeast phenylalanine tRNA. Proc. Tenth FEBS Meet. , 39 , 183–206. [ Google Scholar ]
- Crick F.H.C. (1966) Codon–anticodon pairing: The wobble hypothesis. J. Mol. Biol. , 19 , 548–555. [ PubMed ] [ Google Scholar ]
- Doudna J.A., Cormack, B.P. and Szostak, J.W. (1989) RNA structure, not sequence, determines the 5′ splice-site specificity of a group I intron. Proc. Natl Acad. Sci. USA , 86 , 7402–7406. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ferre-D’Amare A., Zhou, K. and Doudna, J.A. (1998) Crystal structure of a hepatitis delta ribozyme. Nature , 395 , 567–574. [ PubMed ] [ Google Scholar ]
- Gabriel K., Schneider, J. and McClain, W.H. (1996) Functional evidence for indirect recognition of G·U in tRNA Ala by alanyl-tRNA synthetase. Science , 271 , 195–197. [ PubMed ] [ Google Scholar ]
- Gautheret D., Konings, D. and Gutell, R.R. (1995) G·U base pairing motifs in ribosomal RNA. RNA , 1 , 807–814. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Hermann T. and Westhof, E. (1999) Non Watson–Crick base pairs in RNA–protein recognition. Chem. Biol. , 6 , R335–R343. [ PubMed ] [ Google Scholar ]
- Holley R.W., Apgar, J., Everett, G.A., Madison, J.T., Marquisee, M., Merrill, S.H., Penswick, J.R. and Zamir, A. (1965) Structure of ribonucleic acid. Science , 147 , 1462–1465. [ PubMed ] [ Google Scholar ]
- Hou Y.-M. and Schimmel, P. (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature , 333 , 140–145. [ PubMed ] [ Google Scholar ]
- Hur M. and Waring, R.B. (1995) Two group I introns with a C·G basepair at the 5′ splice-site instead of the very highly conserved U·G basepair: is selection post-translational? Nucleic Acids Res. , 23 , 4466–4470. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Illangasekare M. and Yarus, M. (1999) A tiny RNA that catalyzes both aminoacyl-RNA and peptidyl-RNA synthesis. RNA , 5 , 1482–1489. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Jaeger J.A., Turner, D.H. and Zuker, M. (1989) Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA , 86 , 7706–7710. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Khvorova A., Kwak, Y.-G., Tamkun, M., Majerfeld, I. and Yarus, M. (1999) RNAs that bind and change the permeability of phospholipid membranes. Proc. Natl Acad. Sci. USA , 96 , 10649–10654. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Kieft J.S. and Tinoco, I.J. (1997) Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt (III) hexamine. Structure , 5 , 713–721. [ PubMed ] [ Google Scholar ]
- Konforti B.B., Abramowitz, D.L., Duarte, C.M., Karpeisky, A., Beigelman, L. and Pyle, A.M. (1998) Ribozyme catalysis from the major groove of group II intron domain 5. Mol. Cell , 1 , 433–441. [ PubMed ] [ Google Scholar ]
- Ladner J.E., Jack, A., Robertus, J.D., Brown, R.S., Rhodes, D., Clark, B.F.C. and Klug, A. (1975) Structure of yeast phenylalanine transfer RNA at 2.5 Å resolution. Proc. Natl Acad. Sci. USA , 72 , 4414–4418. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Li B., Vilardel, J. and Warner, J.R. (1996) An RNA structure involved in feedback regulation of splicing and translation is critical for biological fitness. Proc. Natl Acad. Sci. USA , 93 , 1596–1600. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Masquida B. and Westhof, E. (2000) On the wobble G·U and related pairs. RNA , 6 , 9–15. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Masquida B., Sauter, C. and Westhof, E. (1999) A sulfate pocket formed by the G·U pairs in the 0.97 Å resolution X-ray structure of a nonameric RNA. RNA , 5 , 1384–1395. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Mathews D.H., Sabina, J., Zuker, M. and Turner, D.H. (1999) Expanded sequence dependence of thermodynamic parameters provides improved prediction of RNA secondary structure. J. Mol. Biol. , 288 , 911–940. [ PubMed ] [ Google Scholar ]
- McClain W.H. and Foss, K. (1988) Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science , 240 , 793–796. [ PubMed ] [ Google Scholar ]
- McClain W.H., Jou, Y.-Y., Bhattacharya, S., Gabriel, K. and Schneider, J. (1999) The reliability of in vivo structure-function analysis of tRNA aminoacylation. J. Mol. Biol. , 290 , 391–409. [ PubMed ] [ Google Scholar ]
- McDowell J.A. and Turner, D.H. (1996) Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC) 2 by two-dimensional NMR and simulated annealing. Biochemistry , 35 , 14077–14089. [ PubMed ] [ Google Scholar ]
- Mueller U., Schübel, H., Sprinzl, M. and Heinemann, U. (1999) Crystal structure of acceptor stem of tRNA Ala from Escherichia coli shows unique G·U wobble base pair at 1.16 Å resolution. RNA , 5 , 670–677. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Musier-Forsyth K. and Schimmel, P. (1992) Functional contacts of a transfer RNA synthetase with 2′-hydroxyl groups in the RNA minor groove. Nature , 357 , 513–515. [ PubMed ] [ Google Scholar ]
- Musier-Forsyth K., Usman, N., Scaringe, S., Doudna, J.A., Green, R. and Schimmel, P. (1991) Specificity for aminoacylation of an RNA helix: An unpaired exocyclic amino group in the minor groove. Science , 253 , 784–786. [ PubMed ] [ Google Scholar ]
- Ott G., Arnold, L. and Limmer, S. (1993) Proton NMR studies of manganese ion binding to tRNA-derived acceptor arm duplexes. Nucleic Acids Res. , 21 , 5859–5864. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Peebles C.L., Zhang, M., Perlman, P.S. and Franzen, J.S. (1995) Catalytically critical nucleotides in domain 5 of a group II intron. Proc. Natl Acad. Sci. USA , 92 , 4422–4426. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Perrotta A.T. and Been, M.D. (1996) Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. , 24 , 1314–1321. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Pyle A.M. (1993) Ribozymes: A distinct class of metalloenzymes. Science , 261 , 709–714. [ PubMed ] [ Google Scholar ]
- Pyle A.-M., Moran, S., Strobel, S.A., Chapman, T., Turner, D.H. and Cech, T.R. (1994) Replacement of the conserved G·U with a G-C pair at the cleavage site of the Tetrahymena ribozyme decreases binding, reactivity and fidelity. Biochemistry , 33 , 13856–13863. [ PubMed ] [ Google Scholar ]
- Quigley G.J. and Rich, A.R. (1975) Structural domains of transfer RNA molecules. Science , 194 , 796–806. [ PubMed ] [ Google Scholar ]
- Ramos A. and Varani, G. (1997) Structure of the acceptor stem of Escherichia coli tRNA Ala : Role of the G3·U70 base pair in synthetase recognition. Nucleic Acids Res. , 25 , 2083–2090. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Rudinger J., Hillenbrandt, R., Sprinzl, M. and Giegé, R. (1996) Antideterminants present in minihelix Sec hinder its recognition by prokaryotic elongation factor Tu. EMBO J. , 15 , 650–657. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ruff M., Krishnaswamy, S., Boeglin, M., Poterszman, A., Mitschler, A., Podjarny, A., Rees, B., Thierry, J.C. and Moras, D. (1991) Class II aminoacyl transfer RNA synthetases: Crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA Asp . Science , 252 , 1682–1689. [ PubMed ] [ Google Scholar ]
- Sprinzl M., Horn, C., Brown, M., Ioudovitch, A. and Steinberg, S. (1996) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. , 26 , 148–153. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Strazewski P., Biala, E., Gabriel, K. and McClain, W.H. (1999) The relationship of thermodynamic stability at a G·U recognition site to tRNA aminoacylation specificity. RNA , 5 , 1490–1494. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Strobel S.A. and Cech, T.R. (1995) Minor groove recognition of the conserved G·U pair at the Tetrahymena ribozyme reactive site. Science , 267 , 675–679. [ PubMed ] [ Google Scholar ]
- Strobel S.A. and Cech, T.R. (1996) Exocyclic amine of the conserved G·U pair at the cleavage site of the Tetrahymena ribozyme contributes to 5′-splice site selection and transition state stabilization. Biochemistry , 35 , 1201–1211. [ PubMed ] [ Google Scholar ]
- Westhof E., Dumas, P. and Moras, D. (1985) Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol. , 184 , 119–145. [ PubMed ] [ Google Scholar ]
How Can Multiple Codons Code For The Same Amino Acid?
Mrna: the starting point of translation, trna: the assembly point of translation, wobble hypothesis: why 20 amino acids have 64 codons, the significance of the wobble hypothesis.
The wobble hypothesis explains that the binding between the 3rd codon base and the 1st anticodon base does not follow canonical Watson-Crick base pairing, allowing multiple codons to code for a single amino acid.
There are 20 different amino acids that can make up a protein (22 if you count the rare amino acids : selenocysteine and pyrrolysine). Each amino acid is formulated with the help of ribosomes, mRNA and tRNA, which together make up the protein-generating machinery. Amino acids are added one after the other in the ribosome, like beads in a necklace. The resulting ‘necklace’ is a polypeptide chain that is then folded into a protein.
Before we uncover how multiple codons can code for a single amino acid, let’s learn a bit more about the players involved.
Recommended Video for you:
Messenger RNA or mRNA is a single-stranded piece of RNA. It is synthesized during a process called transcription, where the information from the DNA is copied or transcribed into an mRNA strand. The double-stranded DNA helix opens up and an mRNA strand identical to the 5’ to 3’ coding strand is produced. Note that ribonucleic acid (RNA) contains the nucleotide uridine (U) in place of thymine (T), which is the only difference between the mRNA and the coding (5’ to 3’) strand of the DNA.
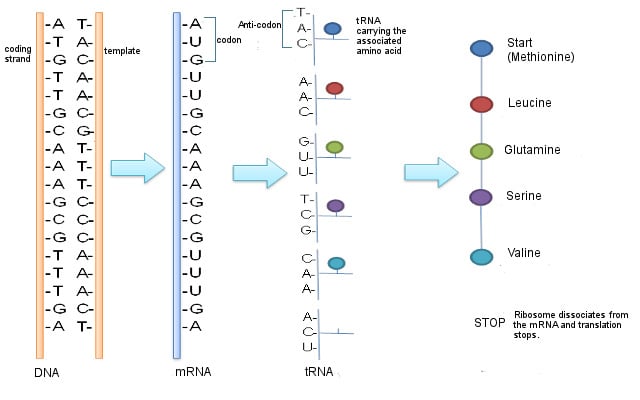
The mRNA then makes its way to the protein-making machinery—the ribosomes. The ribosomes read the information in mRNA to make proteins. This is called translation, like translating instructions to make something.
The information in the mRNA is read three nucleotides at a time in sections called codons . Each codon specifies a certain amino acid.
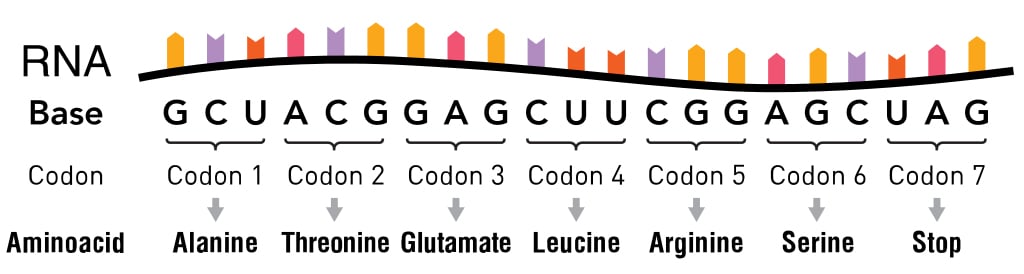
For example, the codon AUG codes for the amino acid methionine (Met). AUG also codes where the protein information starts, called the start codon, which is required to initiate the translation process. Thus, methionine is always the first amino acid in an amino acid chain. The amino acid tryptophan (Trp) is indicated by the codon UGG.
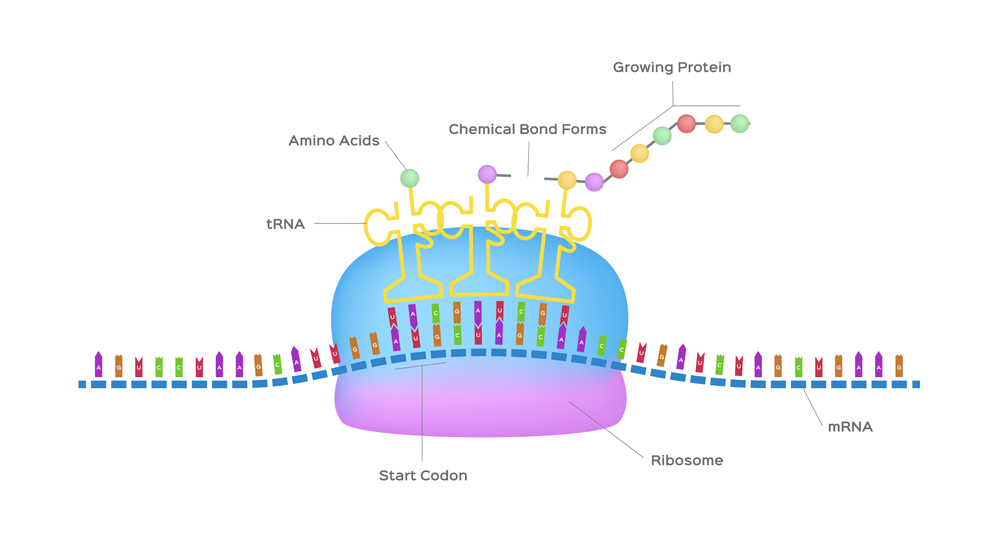
Codons don’t only code for amino acids; they also code for when to terminate the translation process. They are collectively known as stop codons and are UAG, UAA and UGA.
Hence, a codon is like a three-letter password that is required to obtain an amino acid and marks where the instructions to start and stop making the protein are found.
Also Read: What Does RNA Do In A Cell?
The transfer RNA or tRNA is also single-stranded, but is folded, unlike the straight mRNA. It serves as a physical link between the mRNA and the amino acids by interpreting the mRNA and transferring the right amino acids to their place while making the protein. If the translation machinery was a factory, the tRNA would be the factory workers who interpret the instruction manual (the mRNA) to carefully put products in the specified order.
The tRNA is structured in such a way that it has an anticodon loop on one end and an acceptor arm/stem on the opposite end.
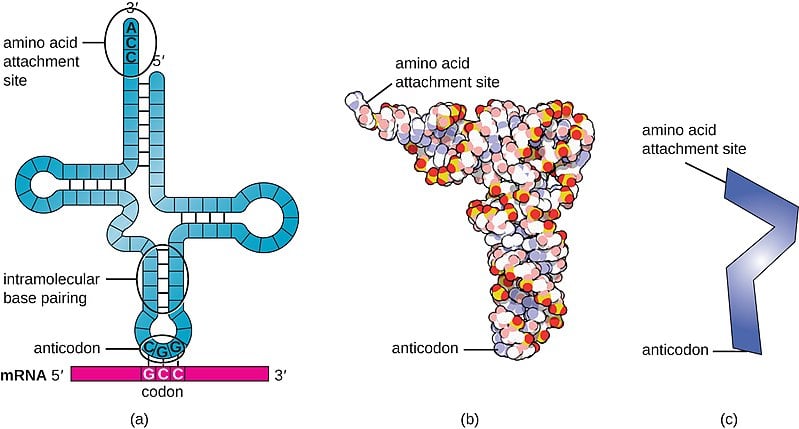
As the name suggests, the anticodon loop is complementary and antiparallel (3’ to 5’) to the mRNA codons. That is, the tRNA consists of the 3 nucleotides that bind to the ones present on the mRNA. Thus, guanine (G) and cytosine (C) bind to each other and adenine (A) and uridine (U) bind to one another.
So, considering the codon for methionine, AUG, it would have a methionine tRNA with an anti-codon UAG.
This type of pairing is known as Watson-Crick base pairing. The pairing is like the attraction between highly specific magnets. Anti-codons on the tRNA ensure that it lines up against the correct codon on the mRNA. Furthermore, as codons are read in the 5’ to 3’ direction, anticodons present on the tRNA are positioned in the 3’ to 5’ direction.
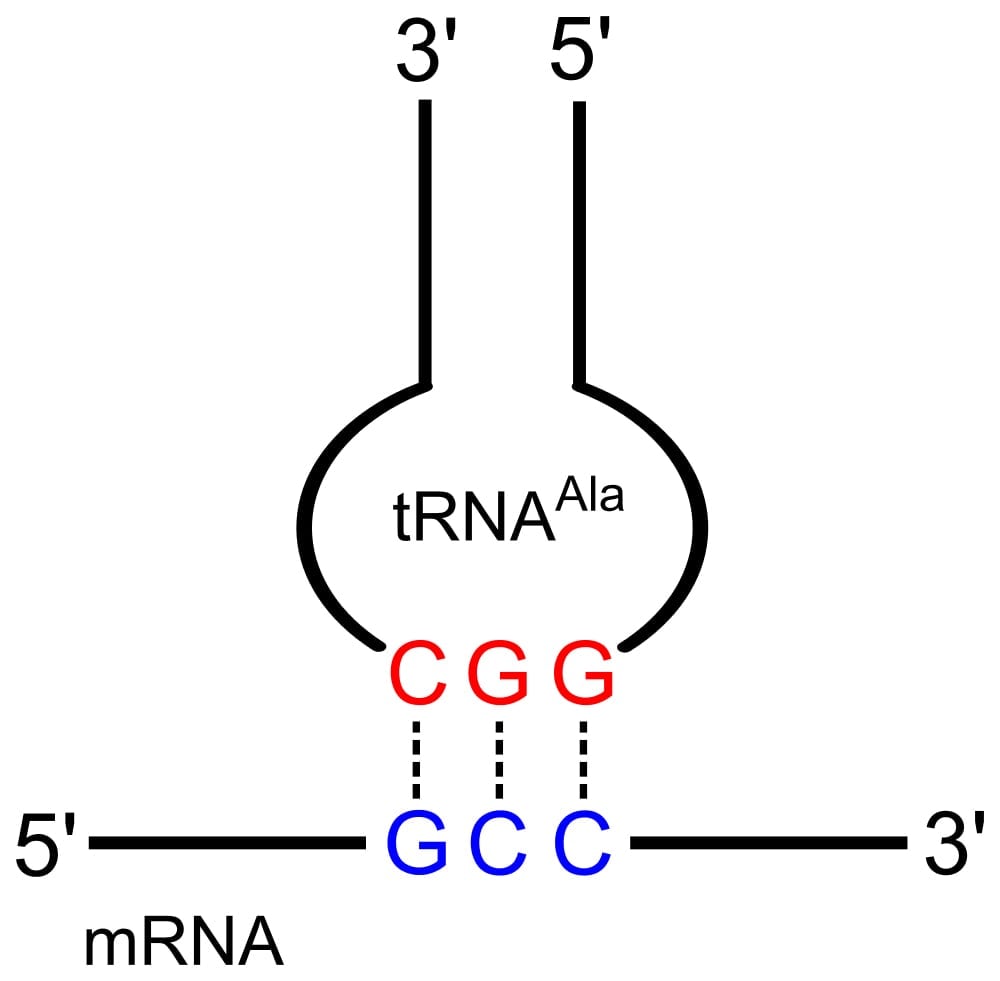
This means that the 1st codon base binds to the 3rd anticodon base and so on. Note that each amino acid has its very own tRNA, which correctly positions it in the polypeptide chain due to the base-pairing between the codon and anticodon.
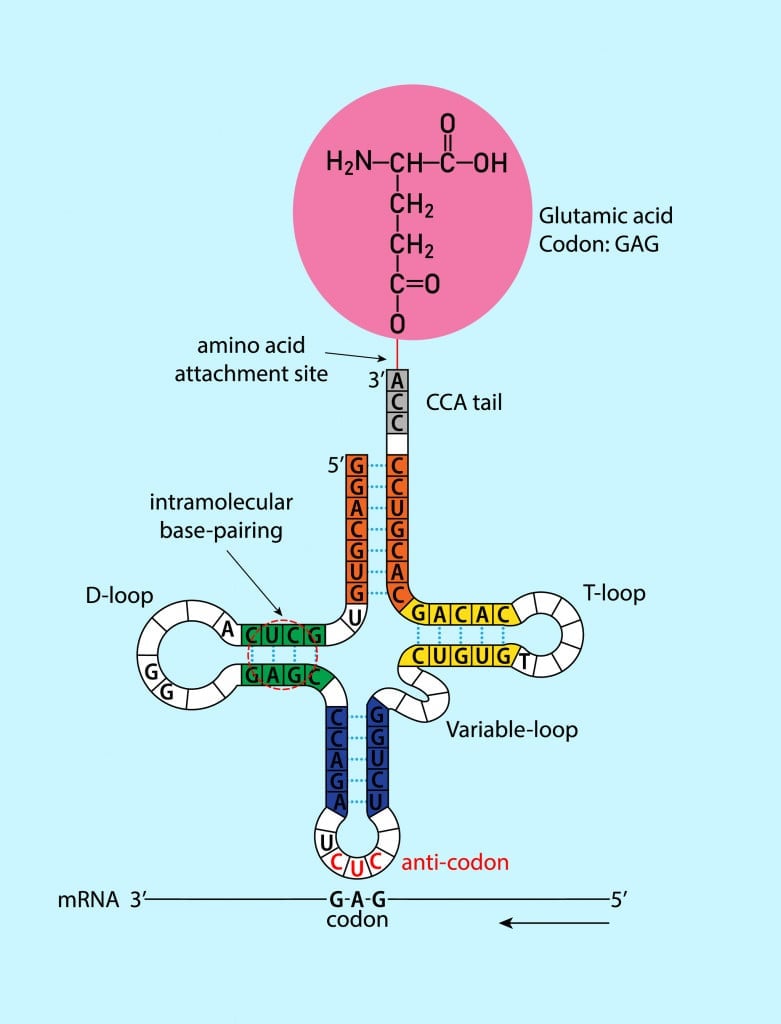
However, like many things in biology, this is also a little more complicated. Each amino acid can be specified by more than one codon. Additionally, the base-pairing rules between the codon and anticodon are not equally binding for all bases. We will learn more about this peculiar phenomenon next!
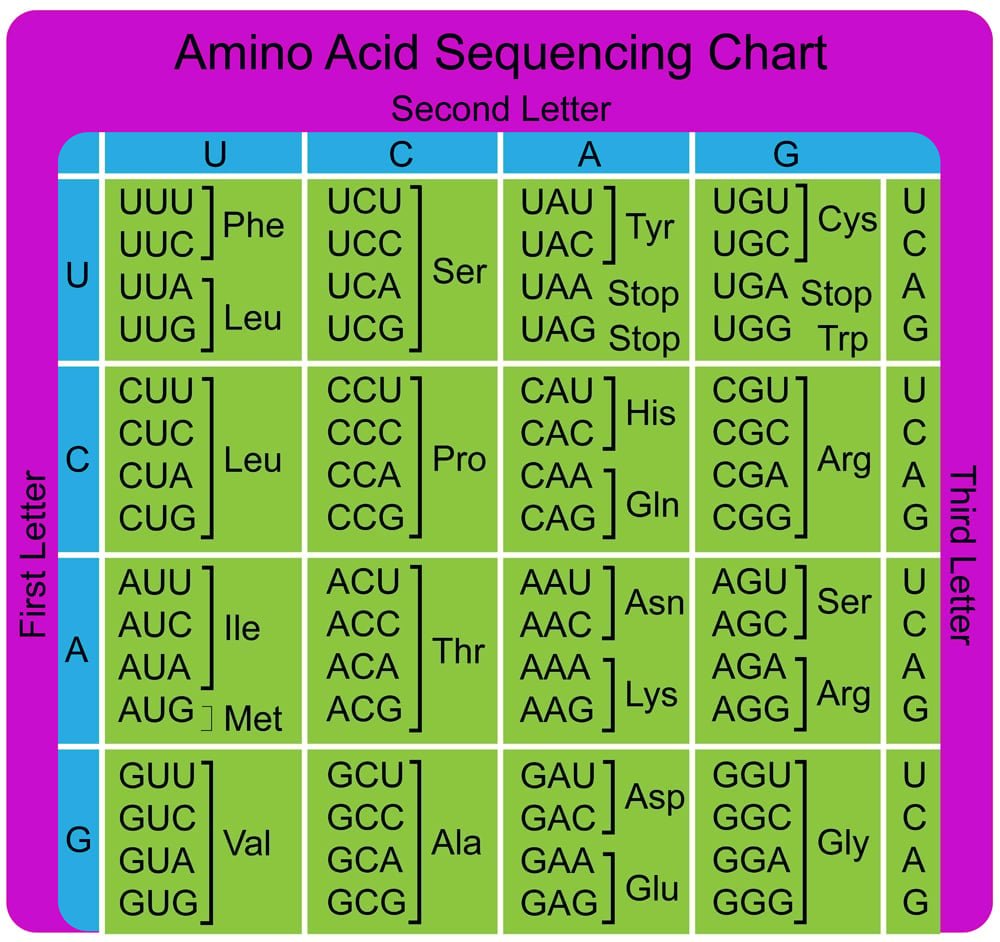
As we know, DNA is made up of 4 nucleotide bases (A, G, T, C). These letters (bases) are read three at a time, which means that there are 64 (4 x 4 x 4) possible combinations of these triplets or codons.
Of the 64 codons, 3 are stop codons, which we mentioned previously. These three stop codons do not code for amino acids and only terminate the process of translation. Therefore, we are left with 61 codons for just 20 amino acids.
The only logical option is that a single amino acid can be coded by multiple codons. But, in theory, 61 different tRNAs would be required for every different codon, yet this isn’t the case. Fewer than 50 codons have been identified in nature.
This observation implies that there might be some leeway between codons and anticodons matching up. Consider the amino acid valine, which is coded by 4 codons: GUU, GUC, GUA, GUG. Notice that only the third base changes, while the first two are the same.
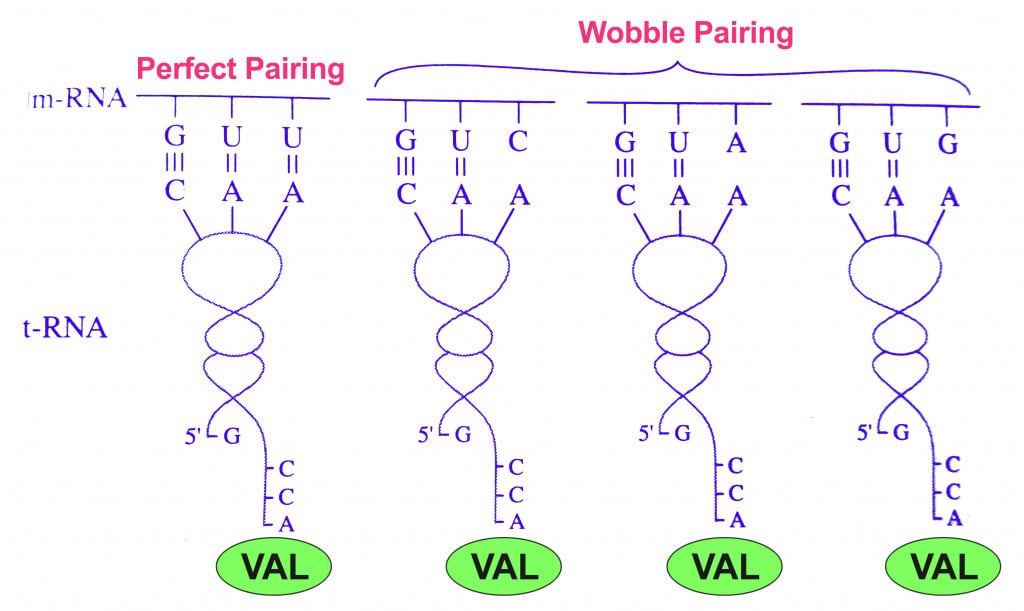
This variance observed only on the 3rd base of the codon led to the ‘ Wobble Hypothesis ’, proposed by Francis Crick (on of the discoverers of DNA structure) in 1966.
The Wobble Hypothesis explained this variance by revealing that the rules of Watson-Crick base-pairing were not followed at the last position of the codon. That is, the bond between the 3rd letter (nucleotide base) and the 1st letter of the anticodon may show unusual bonding (non A-T/ non G-C). Hence, it was allowed to ‘wobble’.
This meant that while the 1st and 2nd bases of the codons strictly adhered to the rules of Watson-Crick base pairing by binding to the 3rd and 2nd base of the anticodon, respectively, non-Watson-Crick base pairing was allowed between the 3rd base of the codon and 1st base of the anticodon. Hence, some tRNAs can recognize many different codons. It also explains the pattern of redundancy in the genetic code (many codons for a single amino acid).
The rules of the Wobble Hypothesis are as follows:
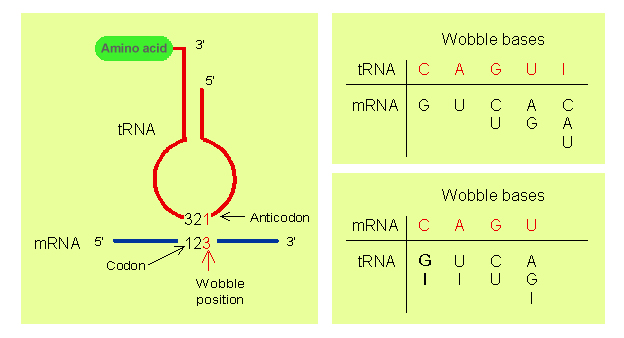
- The first two bases of the codon and the last two bases of the anticodon undergo normal Watson-Crick base pairing. That is, hydrogen bonds are formed between adenine (A) and uridine (U), guanine (G) and cytosine (C), only.
- Less stringent rules of base pairing apply at the remaining position and non-Watson-Crick base pairing may take place. The bases that undergo such pairing are also referred to as wobble base pairs. This allows the anticodon of a single form (amino acid specific) of tRNA to pair with more than one codon in the mRNA.
- When uridine (U) is present in the remaining (1st base of anticodon) position, it can recognize adenine (A) or guanine (G) only.
- When guanine (G) is present in the remaining (1st base of anticodon) position, it can recognize uridine (U) or cytosine (C) only.
- If the modified base inosine (I) is present in the remaining (1st base of anticodon) position, it can recognize uridine (U), cytosine (C) or adenine (A).
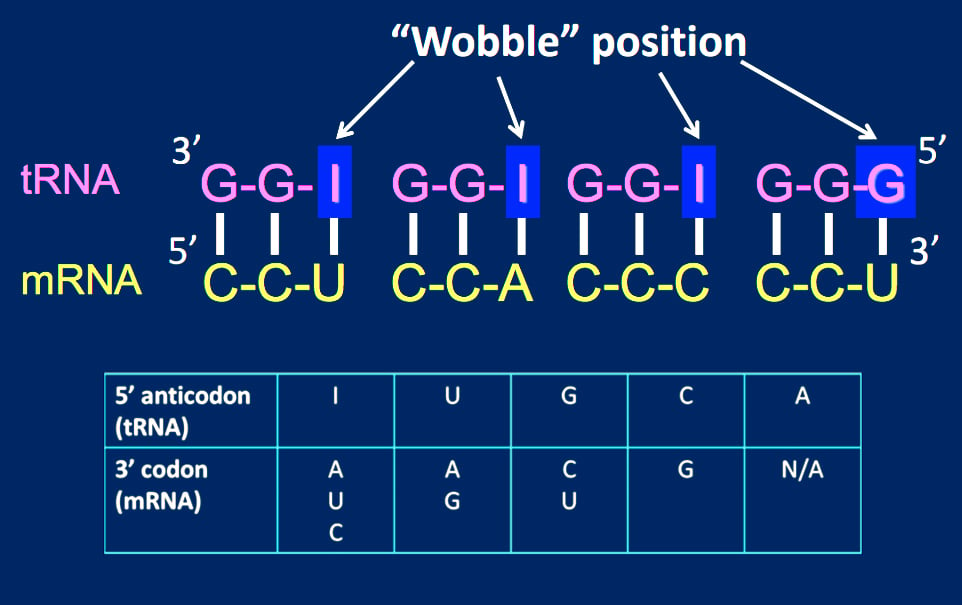
The reason for the wobble base pairing is a slip-up on the part of the ribosome. The ribosome has mechanisms in place to check whether the 1st and 2nd codon bases are complementary to the 3rd and 2nd anticodon bases, respectively. However, the ribosome doesn’t have a mechanism to check whether the 3rd codon base on the mRNA and 1st anticodon base on the tRNA match.
Also Read: What Are The Two Rare Amino Acids?
Slip-up or not, because of the wobble base, there is a lesser chance of mistakes during translation. For example, if the leucine (Leu) codon CUU was misread as CUA, CUG or CUC during transcription, the codon would still be translated as leucine (Leu) during protein synthesis.
Furthermore, protein synthesis is also safeguarded against mutations of a single base. Thus, if the 3rd base in the valine codon GUU mutates and changes to GUC, GUA or GUG, it will still be translated correctly.
Wobble and the degeneracy of the genetic code also reduce the number of tRNAs required by a cell. For instance, for the four different codons of glycine, there are only 3 tRNAs present in E.coli .
So, while it is true that multiple codons can code for a single amino acid, it is the ‘wobble’ observed between the 3rd codon base and the 1st anticodon base that makes this flexibility possible.
- Crick, F. H. C. (1966, August). Codon—anticodon pairing: The wobble hypothesis. Journal of Molecular Biology. Elsevier BV.
- (2014) Intronic and plasmid-derived regions contribute to the large .... National Center for Biotechnology Information
- BB331 Lecture 12 - oregonstate.edu
- How the ribosome decodes a modified 'wobble' position .... The European Synchrotron Radiation Facility
- Wobble Hypothesis - Newcastle University. Newcastle University
- Chapter 5. Genetic Code, Translation, Splicing - Biology. Kenyon College
- Amino Acids Are Encoded by Groups of Three Bases Starting from a Fixed Point - Biochemistry - NCBI Bookshelf - www.ncbi.nlm.nih.gov

Saloni Hombalkar has earned her Bachelor’s degree in Biomedical Sciences from SDSOS, NMIMS-Mumbai. She is an enthusiastic science communicator and hopes to share her passion for biology with as many people as possible. Apart from Biology, she enjoys discussing Marvel, learning to code, reading YA fiction and dramatic karaoke sessions.

What Is A Deletion Mutation?

Is 98% Of Our DNA Really Junk?
What is a mutation what are the different types of mutations.

What Is DNA And How Does It Work?

What Is A Phosphodiester Bond?

Does Human DNA Change With Time?

Higgs Boson (The God Particle) and Higgs Field Explained in Simple Words

Quantum Entanglement: Explained in REALLY SIMPLE Words

What are Mutations and what are the different types of Mutations?

Resonance (Chemistry) Explained in Simple Words with Examples

Isotopes Explained in Simple Words with Real-life Examples

What is Evolution: A REALLY SIMPLE and Brief Explanation
This page has been archived and is no longer updated
Further Exploration
Concept Links for further exploration

- Gene Inheritance and Transmission
- Gene Expression and Regulation
- Nucleic Acid Structure and Function
- Chromosomes and Cytogenetics
- Evolutionary Genetics
- Population and Quantitative Genetics
- Genes and Disease
- Genetics and Society
- Cell Origins and Metabolism
- Proteins and Gene Expression
- Subcellular Compartments
- Cell Communication
- Cell Cycle and Cell Division
© 2014 Nature Education
- Press Room |
- Terms of Use |
- Privacy Notice |


IMAGES
VIDEO
COMMENTS
The wobble hypothesis states that the base at 5' end of the anticodon is not spatially confined as the other two bases allowing it to form hydrogen bonds with any of several bases located at the 3' end of a codon. ... for example, if the Leu codon CUU were misread CUC or CUA or CUG during transcription of mRNA, the codon would still be ...
Examples of Wobble Hypothesis. Here are some examples of the wobble hypothesis in action: Arginine: The amino acid arginine is coded by six different codons: CGU, CGC, CGA, CGG, AGA, and AGG. However, no six different tRNA molecules correspond to each of these codons. Instead, one tRNA molecule with the anticodon 3′-CCU-5′ can recognize the ...
The purine, inosine, is a wobble nucleotide and is similar to guanine which normally pairs with A, U and C. For example a glycine-tRNA with anticodon 5′-ICC-3′ will pair with glycine codons GGU, GGC, GGA and GGG (Fig 7.2). ... The U at the wobble position will be able to pair with an adenine or a guanine. According to Wobble hypothesis ...
The Wobble Hypothesis states that position 34 inosine may base pair with uridine, cytidine, and adenosine. ... or the recognition of multiple codons can be made feasible with various chemical moieties introduced onto the wobble base. 5 There are several examples of both expansion as well as restriction of codon recognition which are presented ...
Wobble base pairs for inosine and guanine. A wobble base pair is a pairing between two nucleotides in RNA molecules that does not follow Watson-Crick base pair rules. The four main wobble base pairs are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C).In order to maintain consistency of nucleic acid nomenclature, "I" is used for ...
For example Inosine (I) can pair with A, C or U. this base is called Wobble base or fluctuating base Wobble occurs at position 1 of the anti-codon and position 3 of the codon. The wobble hypothesis states that the third position (3') of the codon on mRNA and the first position (5') of the anticodon on tRNA are bound less tightly than the ...
The Wobble Hypothesis, proposed by Francis Crick in 1966, explains the degeneracy of the genetic code. The hypothesis attributes this degeneracy to imprecise pairing between the third base of the codon and the first base of the anticodon on tRNA. ... For example, G may pair with T or U. The wobble hypothesis is important in translating the ...
The Wobble Hypothesis explains why multiple codons can code for a single amino acid. One tRNA molecule (with one amino acid attached) can recognize and bind ...
The Wobble Hypothesis •The first two bases of the codon make normal (canonical) H- bond pairs with the 2nd and 3rd bases of the anticodon •At the remaining position, less stringent rules apply and non- canonical pairing may occur •The rules: first base U can recognize A or G, first base G can recognize U or C, and first base I can recognize U, C or A (I
F.H.C. Crick, in 1965 proposed a hypothesis called Wobble hypothesis to explain this phenomenon. He discovered that if U is present at first position of anticodon, it can pair with either A or G at the third position of codon. Similar is the case with G and I (I = inosine is a modified base in tRNA) found in anticodon. The pairing relationships ...
Wobble base pairs occur frequently in RNA secondary structure and are important for proper translation of the genetic code. In 1966, Francis Crick proposed the Wobble hypothesis to account for the fact that most organisms do not seem to have as many tRNA molecules as would be required for complete translation of the genetic code. There are 64 ...
According to the wobble hypothesis , G34 base-pairs with C and U as the third nucleoside of the codon [denoted C(III) and U(III)], whereas C34 only base-pairs with G(III). Uridine as the wobble nucleoside cannot interact with a pyrimidine in the mRNA, since two pyrimidines are too "short" to form a base pair.
wobble hypothesis A theory proposed to explain the partial degeneracy of the genetic code in that some t-RNA molecules can recognize more than one codon.It is proposed that the first 2 bases in the codon and anticodon will form complementary pairs in the normal antiparallel fashion. However, a degree of steric freedom or 'wobble' is allowed in the base pairing at the third position.
Wobble base pairing enables the decoding of two or more codons by the same tRNA. Certain modified bases, for example inosine (I) can extend or restrict the degree of flexibility in the range of mRNA:tRNA interactions. Codons that are decoded by wobble base interactions are processed at a slower rate in the ribosome.
The 'modified wobble hypothesis' suggested that specific tRNA base modifications evolved to ... shift of nucleobases towards minor or major groove, characteristic of, for example, wobble G·U ...
Search for: 'wobble hypothesis' in Oxford Reference ». A theory to explain the partial degeneracy of the genetic code due to the fact that some t-RNA molecules can recognize more than one codon. The theory proposes that the first two bases in the codon and anticodon will form complementary pairs in the normal antiparallel fashion.
The G·U wobble base pair is a fundamental unit of RNA secondary structure that is present in nearly every class of RNA from organisms of all three phylogenetic domains. ... For example, the wobble pair at the third position of the acceptor helix of tRNA ... Codon-anticodon pairing: The wobble hypothesis. J. Mol. Biol., 19, 548-555. [Google ...
The wobble hypothesis explains that the binding between the 3rd codon base and the 1st anticodon base does not follow canonical Watson-Crick base pairing, allowing multiple codons to code for a single amino acid. ... For example, the codon AUG codes for the amino acid methionine (Met). AUG also codes where the protein information starts, called ...
Wobble and Superwobble. In most cases, more than one triplet codon can specify an amino acid—at one extreme, leucine can be encoded by any of six nucleotide triplets. Degenerate codons tend to vary at the third position, which was the basis for Francis Crick's wobble hypothesis: Each codon must be recognized by its cognate transfer RNA (tRNA ...
Codon—anticodon pairing: The wobble hypothesis. It is suggested that while the standard base pairs may be used rather strictly in the first two positions of the triplet, there may be some wobble in the pairing of the third base. This hypothesis is explored systematically, and it is shown that such a wobble could explain the general nature of ...
Wobble Hypothesis. the hypothesis that some tRNA molecules can pair with more than one mRNA codon, tolerating some variations in the third base, as long as the first and second bases are correctly matched. C is base in anticodon at 5'. G is base in codon at 3'. U is base in anticodon at 5'. A or G is base in codon at 3'.
wobble. Describes the redundancy in the genetic code such that the same amino acid may be encoded by multiple codons.
The Wobble Hypothesis states that position 34 inosine may base pair with uridine, cytidine, and adenosine. The ability of inosine at the wobble position to promote the reading of multiple codons, in some cases, proves essential to survival. ... As an example, isoleucine shares a codon box with methionine where AUA, AUC, and AUU code for ...