- Open access
- Published: 30 December 2022

A one-year genomic investigation of Escherichia coli epidemiology and nosocomial spread at a large US healthcare network
- Emma G. Mills 1 na1 ,
- Melissa J. Martin 1 na1 ,
- Ting L. Luo 1 ,
- Ana C. Ong 1 ,
- Rosslyn Maybank 1 ,
- Brendan W. Corey 1 ,
- Casey Harless 1 ,
- Lan N. Preston 1 ,
- Joshua A. Rosado-Mendez 1 ,
- Scott B. Preston 2 ,
- Yoon I. Kwak 1 ,
- Michael G. Backlund 2 ,
- Jason W. Bennett 1 ,
- Patrick T. Mc Gann 1 &
- Francois Lebreton ORCID: orcid.org/0000-0002-7157-5026 1
Genome Medicine volume 14 , Article number: 147 ( 2022 ) Cite this article
5297 Accesses
15 Altmetric
Metrics details
Extra-intestinal pathogenic Escherichia coli (ExPEC) are a leading cause of bloodstream and urinary tract infections worldwide. Over the last two decades, increased rates of antibiotic resistance in E. coli have been reported, further complicating treatment. Worryingly, specific lineages expressing extended-spectrum β -lactamases (ESBLs) and fluoroquinolone resistance have proliferated and are now considered a serious threat. Obtaining contemporary information on the epidemiology and prevalence of these circulating lineages is critical for containing their spread globally and within the clinic.
Whole-genome sequencing (WGS), phylogenetic analysis, and antibiotic susceptibility testing were performed for a complete set of 2075 E. coli clinical isolates collected from 1776 patients at a large tertiary healthcare network in the USA between October 2019 and September 2020.
The isolates represented two main phylogenetic groups, B2 and D, with six lineages accounting for 53% of strains: ST-69, ST-73, ST-95, ST-131, ST-127, and ST-1193. Twenty-seven percent of the primary isolates were multidrug resistant (MDR) and 5% carried an ESBL gene. Importantly, 74% of the ESBL -E.coli were co-resistant to fluoroquinolones and mostly belonged to pandemic ST-131 and emerging ST-1193. SNP-based detection of possible outbreaks identified 95 potential transmission clusters totaling 258 isolates (12% of the whole population) from ≥ 2 patients. While the proportion of MDR isolates was enriched in the set of putative transmission isolates compared to sporadic infections (35 vs 27%, p = 0.007), a large fraction (61%) of the predicted outbreaks (including the largest cluster grouping isolates from 12 patients) were caused by the transmission of non-MDR clones.
By coupling in-depth genomic characterization with a complete sampling of clinical isolates for a full year, this study provides a rare and contemporary survey on the epidemiology and spread of E. coli in a large US healthcare network. While surveillance and infection control efforts often focus on ESBL and MDR lineages, our findings reveal that non-MDR isolates represent a large burden of infections, including those of predicted nosocomial origins. This increased awareness is key for implementing effective WGS-based surveillance as a routine technology for infection control.
Extra-intestinal pathogenic Escherichia coli (ExPEC) are a leading cause of healthcare-associated urinary tract and bloodstream infections [ 1 , 2 ]. Diseases caused by multidrug-resistant (MDR) strains are associated with poor patient outcomes, including high morbidity and mortality, and higher healthcare costs [ 3 , 4 , 5 ]. In recent years, resistance to commonly prescribed antibiotics has increased in E. coli infections in the USA [e.g., 1.2 to 25% prevalence of fluoroquinolone resistance in the past 15 years [ 6 , 7 ]] and internationally [ 1 , 3 , 8 , 9 ]. Importantly, resistance to 3rd- and 4th-generation cephalosporins, due to the acquisition and horizontal spread of extended-spectrum β -lactamase (ESBL) genes, has increased in both healthcare and community settings [ 10 ]. This alarming rise prompted the US Centers for Disease Control and Prevention to identify the ESBL-producing E. coli as a serious threat and urging increased surveillance efforts [ 11 ].
Previous molecular studies have separated E. coli into phylogenetic groups, including A, B1, B2, C, D, E, and F, with ExPEC (and consequently the specialized uropathogenic [UPEC] pathotype) largely belonging to phylogroups B2 and D [ 12 , 13 ]. Multilocus sequence typing (MLST) provides further characterization of E. coli lineages and has led to the identification of specific, globally distributed sequence types (STs). For example, the ST-131 ExPEC lineage is widely distributed and associated with the emergence of fluoroquinolone resistance and frequent carriage of plasmid-bound ESBL genes [ 12 , 14 , 15 , 16 , 17 ]. Besides resistances, recent studies suggest that the acquisition of virulence-associated genes also plays an integral role in the success and global emergence of ST-131 and other ExPEC lineages. These include a plethora of both structural (e.g., fimbriae, pili, curli, flagella) and secreted (e.g., toxins, iron-acquisition systems) virulence factors often enriched in non-MDR, UPEC lineages (e.g., ST-73, 95, and 127) [ 18 , 19 , 20 ].
The recent positioning of whole-genome sequencing (WGS) as a near-routine technology is creating a revolution in infection control and allows for targeted interventions to reduce the burden of healthcare-associated infections (HAIs). Such effort requires an understanding of the frequency of nosocomial transmission caused not only by MDR epidemic clones, but also by the more ubiquitous non-MDR lineages. While the latter are responsible for most E. coli infections, very few genome-based studies have examined their role in nosocomial transmission. Instead, most investigations have been performed on small cohorts, often limited to ESBL-producing isolates, which likely underrepresents the extent of E. coli nosocomial transmission events [ 21 ].
Here, we retrospectively genome-sequenced and analyzed a complete set of 2075 E. coli clinical isolates collected from 1776 patients over a 12-month period from a large military healthcare network in the Northeast United States. Genome-based detection of possible outbreak clusters revealed extensive roles for non-MDR lineages in suspected nosocomial transmissions, while in-depth phylogenetic, genotypic, and phenotypic characterization revealed a detailed picture of the epidemiology, population structure, and prevalence of resistances in E. coli in this region.
Isolation and phenotypic characterization of E. coli collection
A total of 2075 E. coli isolates (including serial isolates from the same patient) cultured from all clinical specimens of 1776 patients receiving care in the National Capitol Medical Region healthcare network between October 2019 and September 2020 were collected. Of note, no stool isolates were collected as these samples are not routinely sent for culture in the microbiology lab of this hospital and are instead analyzed by molecular and/or antigen diagnostic procedures. Antibiotic susceptibility testing (AST) was performed in a College of American Pathologists (CAP)-certified laboratory using the BD Phoenix (panel NMIC/ID304; BD Diagnostics), which encompasses 18 antibiotics from 11 different antibiotic classes. Where necessary, MICs were determined in triplicate using broth microdilution using Clinical and Laboratory Standards Institute (CLSI) guidelines [ 22 ]. Breakpoints were interpreted using CLSI guidelines (2018), with cefazolin MICs interpreted using breakpoints for complicated UTI/systemic infection [ 22 ]. Isolates with breakpoints interpreted as I or R were designated non-susceptible. To accurately calculate the prevalence of resistances in the population, a subset of 1828 primary isolates (first isolate of each ST per patient) was specifically used (Table 1 ).
Whole-genome sequencing
DNA extraction and WGS were performed as previously described [ 23 ]. In brief, genomes were generated for all 2075 isolates using an Illumina MiSeq platform with a 2×300 nt paired-end protocol or a NextSeq-500 platform with a 2×150 nt paired-end protocol. Libraries were prepared using the Kapa HyperPlus kit (Roche Diagnostics) and quantified using the Kapa library quantification kit Illumina/Bio-Rad iCycler (Roche Diagnostics) on a CFX96 real-time cycler (Bio-Rad). De novo assemblies were obtained using Newbler v2.7 (Roche Diagnostics). Minimum thresholds for contig size and coverage were set at 200 bp and 49.5+, respectively. Assembled sequences were annotated using Prokka v1.14.6 [ 24 ].
Bioinformatic analysis
Species identification was determined using Kraken2 (v2.0.8-β) [ 25 ] and E. coli phylogenetic groups were identified using EzClermont v0.6.3 [ 26 , 27 ]. In silico ST detection was identified for all isolates using the Achtman MLST scheme through software [ 28 ]. This tool uses the PubMLST website [ 29 ] developed by Keith Jolley and sited at the University of Oxford. Novel ST was assigned using the MLST sequence archive at EnteroBase [ 30 ]. Serotyping and fimH typing were performed using the TORMES pipeline v1.3.0 [ 31 ] with the SerotypeFinder O-typing database [ 32 ] and FimTyper [ 33 ], respectively. Antimicrobial resistance genetic determinants were annotated using AMRFinderPlus [ 34 ] and ARIBA [ 35 ]. Plasmid replicons [ 36 ] and virulence-associated genes (based on the E. coli virulence-associated gene databases EcVGDB and VFDB [ 37 , 38 ]) were identified using ABRicate [ 39 ].
Phylogenetic analysis
For the phylogeny of the diverse set of 123 ESBL- E. coli , the annotated [Prokka v1.14.6 [ 24 ]] assemblies were used as input for Roary v3.13.0 [ 40 ] and a SNP-based alignment of 2698 core genes was generated. For the phylogeny of the clonal set of 275 ST-131 E. coli, SNP calling was performed with Snippy v.4.4.5 [ 41 ] using error correction [Pilon v1.23 [ 42 ]] and the annotated genome of ST-131 E. coli EC958 (accession no. GCA_000285655.3) as a reference. For both approaches, recombination was filtered from the alignments using Gubbins v2.4.1 [ 43 ] and a maximum-likelihood tree was generated with RAxML v8.2.12 [ 44 ] using the GTR+G (50 parsimony, 50 random) model and 100 random bootstrap replicates. Trees were imported in iTOL v.5.5 [ 45 ] for visualization with metadata.
Finally, for the ST-131 phylogenetic analysis, clade designations (A, B, and C) were generally characterized by the carriage of type 1 fimbriae adhesion fimH alleles ( fimH 41, fimH 22, and fimH 30, respectively) and subclades C0, C1, and C2 based on SNP typing of genetic markers gyrA , parC , and ybbW genetic markers, as previously described [ 16 , 46 ]. The G273A SNP in ybbW (subclade C2-specific allele) was identified using an individual gene alignment produced by Roary v3.13.0 [ 40 ].
Nosocomial transmission analysis
Detection of clusters of transmission was performed in two stages. First, cgMLST allele assignment and minimum spanning tree generation were performed with SeqSphere+ [ 47 ] using the E. coli cgMLST scheme developed by Zhou et al. [ 30 ]. The distance matrix from SeqSphere+ consisted of the pairwise allelic differences between all 2075 E. coli isolates . Using a threshold of ≤10 allelic differences, a level previously identified as indicative of potential E. coli transmission [ 48 ], 105 putative clusters of transmission were identified and comprised isolates from 2 distinct patients or more (Additional file 1 : Table S1). Clusters of serial isolates from single patients were removed. Second, to further investigate these putative clusters, an internal reference genome (first isolate temporally) was picked and whole-genome SNP analysis was individually performed for the 105 clusters. Using a 17 SNP cutoff, a threshold previously identified between patient pairs sharing strong epidemiological links [ 9 ], 95 of the 105 original clusters were confirmed and were further analyzed in this report. To determine the prevalence of MDR isolates in the clusters, primary MDR cluster isolates ( n =228) were used (serial isolates from the same patient and same MDR or non-MDR designation were removed) (Table 1 ).
Isolate collection and population structure
Between October 2019 and September 2020, a set of 2075 E. coli were collected from all 1776 patients who received care within the National Capitol Region healthcare network (located on the East coast of the USA) (Additional file 1 : Table S1). While obtained from 21 facilities, the majority (59%) of the isolates originated from a single, large tertiary care hospital that also served as the central microbiology hub for the remaining 20 facilities. This sampling represents >99% of all E. coli cultured from clinical specimens at the central microbiology laboratory during this 1-year period. Isolates were primarily obtained from urine (93%), followed by bloodstream infections (2%), wound infections (2%), and perirectal swabs (1%). A small number of isolates were cultured from fluid (.07%), tissue (.04%), and respiratory (.01%) cultures (Additional file 1 : Table S1).
WGS and cgMLST analysis revealed a diverse population that resolved into 5 main E. coli phylogenetic groups (Fig. 1 A, Additional file 1 : Table S1), with B2 and D the most represented (71% and 13%, respectively). Molecular typing by in silico MLST indicated the population was composed of 247 STs with 53% belonging to 6 known, globally prevalent STs. These include the epidemic lineages ST-131 ( n = 275), ST-73 ( n = 224), ST-95 ( n = 215), ST-127 ( n = 138), and the emerging ST-1193 ( n = 112) all within phylogroup B2 [ 49 ]. Epidemic lineage ST-69 ( n = 142) was the sole exception, belonging to phylogroup D. Notably, 133 (53%) STs were each found in isolate(s) from single patients and only 52 of 1776 patients carried strains with multiple STs (Additional file 1 : Table S1).
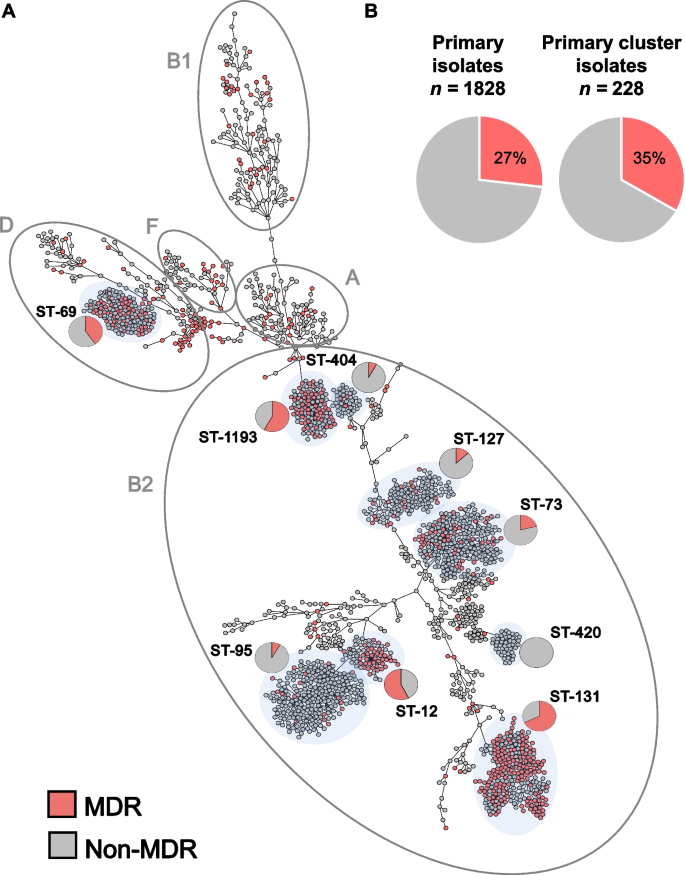
Population structure of a complete collection of E. coli clinical isolates for a 1-year period at a US hospital. A cgMLST-based minimum spanning tree of 2075 E. coli isolates. Isolates belonging to the main phylogenetic groups observed in this study are circled and labeled. The dominant STs are shaded in light gray and the proportion of MDR (red) and non-MDR (gray) isolates within specific STs is indicated by pie charts. B Pie charts indicate the prevalence of MDR (red) primary isolates (27%) was similar to the prevalence of MDR isolates in primary isolates predicted to be part of clusters of transmission (35%) (Table 1 )
Diversity of antibiotic susceptibility profiles
Comprehensive AST was performed on all isolates using 18 antibiotics from 11 different classes (Fig. 2 , Additional file 2 : Table S2). For an accurate determination of the prevalence of resistances in this E. coli population, removal of serial isolates (same ST per patient) resulted in a collection of 1828 primary isolates (Table 1 ). From these, the highest prevalence of non-susceptibility was to ampicillin (41%), followed by tetracycline (23%), trimethoprim/sulfamethoxazole (20%), and fluoroquinolones (15% to ciprofloxacin). In contrast, all isolates were susceptible to amikacin, 5% of E. coli were non-susceptible to third- and fourth-generation cephalosporins and <1% ( n = 14) showed non-susceptibility to a carbapenem. Of the latter, 6 were resistant to imipenem only (MIC = 2), 5 were resistant to ertapenem only (MIC > 0.5 ml/l), 3 were resistant to ertapenem and imipenem or meropenem, and none carried a carbapenemase (Fig. 2 , Table 1 and Additional file 2 : Table S2).
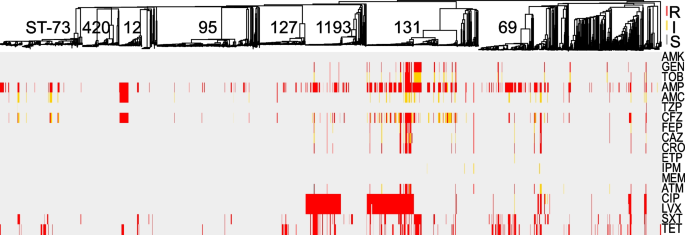
Comprehensive phenotypic antibiotic susceptibility testing of all E. coli isolates to 18 antibiotics from 11 classes tested in this study. Breakpoints were interpreted using CLSI guidelines and S (susceptible), I (intermediate), and R (resistant) classifications are labeled for each antibiotic/isolate: red, yellow, and gray, respectively. Interpretations are mapped onto the MST from Fig. 1 . AMK amikacin, GEN gentamicin, TOB tobramycin, AMP ampicillin, AMC amoxicillin-clavulanic acid, TZP piperacillin-tazobactam, CFZ cefazolin, FEP cefepime, CAZ ceftazidime, CRO ceftriaxone, ETP ertapenem, IPM imipenem, MEM meropenem, ATM aztreonam, CIP ciprofloxacin, LVX levofloxacin, SXT trimethoprim-sulfamethoxazole, TET tetracycline
Distinct lineages of E. coli were enriched for phenotypic resistance to various classes of antibiotics: (i) 50% of ST-12 were non-susceptible to amoxicillin/clavulanate (vs. 13% across all isolates, p < 0.001 by Fisher exact test), (ii) ST-131 accounted for 59% of isolates non-susceptible to gentamicin and tobramycin (vs. 6% for all, p < 0.001), and (iii) ST-131 and ST-1193 alone represented 72% of all isolates with resistance to the fluoroquinolones (Fig. 2 , Additional file 2 : Table S2).
Overall, 27% of the primary isolates were classified as multidrug resistant (MDR) as defined by Magiorkas et al. (i.e., non-susceptible to at least one agent in ≥3 antibiotic categories) [ 50 ] (Fig. 1 A and Additional file 2 : Table S2), though the prevalence of MDR varied significantly among the distinct, most frequent E. coli lineages. For example, while lineages ST-131, ST-1193, and ST-69 were significantly enriched in MDR isolates (64%, 55%, and 43%, respectively, with p -values < 0.01 by Fisher exact test), lineages ST-73, ST-127, and ST-95 largely comprised non-MDR isolates (21%, 13%, and 7%, respectively, with p -values < 0.03) (Fig. 1 A).
Genomic characterization of ESBL-carrying E. coli
During the study period, 123 ESBL-producing E. coli were identified from 90 unique patients and all were classified as MDR (Additional file 3 : Table S3). Interestingly, 22% were cultured from non-urinary sites, a significant divergence from the overall population (7%, p <0.05). Phylogenomic analysis of all ESBL- E. coli isolates indicated ESBL producers were diverse and belonged to 26 STs, including prevalent lineages [ST-131 (from 36 patients), ST-1193 (7 patients), ST-69 (5 patients)], less common lineages in our dataset [ST-38 (11 patients), ST-10 (4 patients)], and rarer ESBL-carrying lineages [ST-44 [ 51 ], ST-256, and ST-636 [ 52 ] each represented by 2 patients each]. As a result, an overrepresentation of ST-131 and ST-1193, which have fluoroquinolone resistance rates of 52% and 100%, respectively, 74% of ESBL-producers were non-susceptible to fluoroquinolones (compared to 17% overall, p < 0.01) (Fig. 3 ). The most represented ESBL genes were bla CTX-M-15 (59%) and bla CTX-M-27 (22%). Furthermore, bla CTX-M-14 was carried by 14% of the isolates including eight ST-38 isolates from 5 patients without an identified plasmid replicon. While carriage on a plasmid with an unknown replicon cannot be ruled out, chromosomal carriage of bla CTX-M-14 has previously been described for strains of this ST collected from Mongolian birds [ 53 ]. bla CTX-M-55 was observed in 3 isolates and bla CTX-M-24 and bla TEM-19 were observed once in distinct lineages (ST-354 and ST-131, respectively) (Fig. 3 ). Notably, the first description of ST-1193 harboring a bla CTX-M-64 allele was observed in a singular isolate (Fig. 3 ). Nine plasmid replicon types regularly associated with ESBL carriage [ 54 , 55 ] were identified with varying prevalence, from ≥10 to 76% (Fig. 3 ).
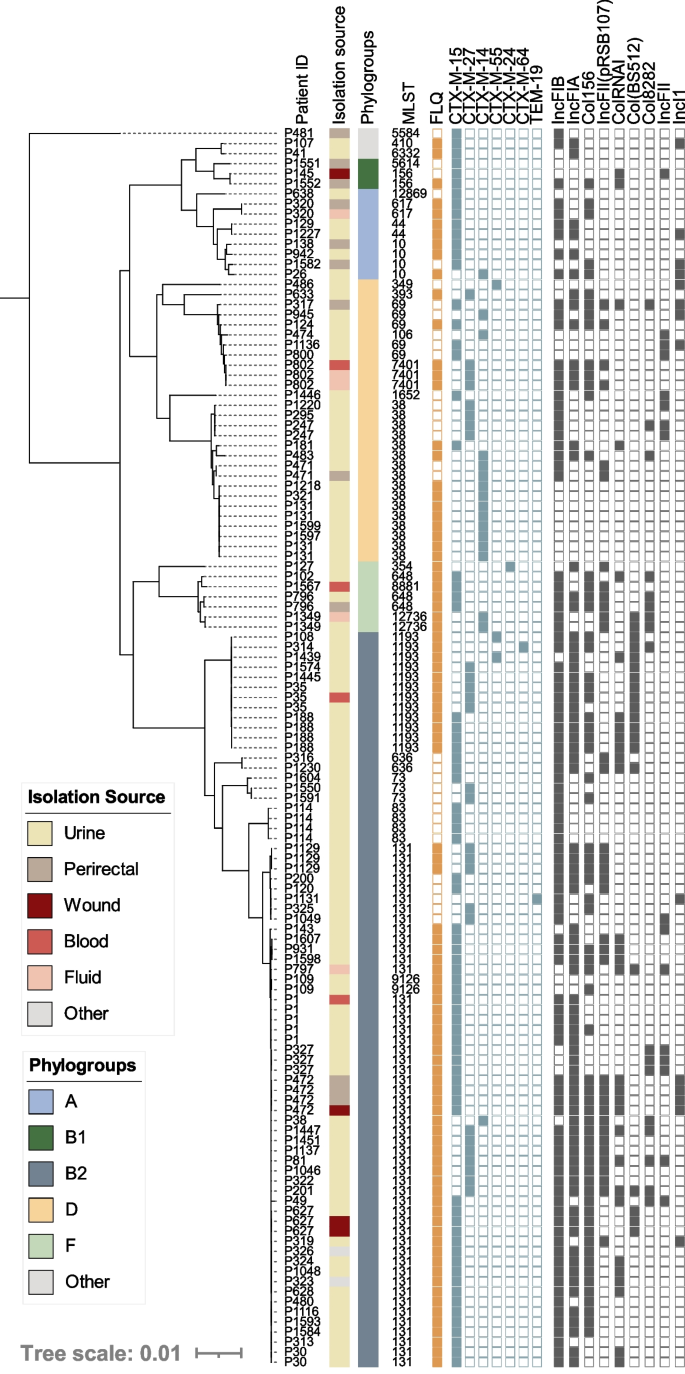
Core genome phylogeny of all ESBL-carrying E. coli isolates in our dataset ( n =123). Patient numbers are listed to identify serial isolates. Isolation source and phylogroups are color coded, indicated by the corresponding legends. Fluoroquinolone (FLQ) (ciprofloxacin and/or levofloxacin) non-susceptibility is indicated by a closed orange square, the presences of unique ESBL alleles are shown in a closed blue square, and plasmid replicon families identified with prevalence ≥10% are indicated by a gray closed square. Two novel ESBL-producing STs were characterized: ST-12869 and ST-12736
Outbreak detection reveals the role of non-MDR E. coli in nosocomial transmission
Prediction of possible clusters of transmission was performed in two steps: cgMLST followed by SNP analysis (Table 1 ). This filtering stringently confirmed 95 clusters (from 105 identified by cgMLST) comprising 258 isolates from 227 patients (Table 1 ). A total of 26 STs were represented and 61% of the clusters (58 out of 95) were caused by a non-MDR clone (Fig. 4 A, B and Additional file 1 : Table S1). At the isolate level, the proportion of primary MDR (35%) isolates from potential outbreaks clusters was slightly increased compared to primary non-cluster isolates (27%, p = 0.007 by Fisher exact test) while the proportion of ESBL producers remained comparable (6%) (Fig. 1 B) (Table 1 ). At the lineage level, the largest number of outbreak clusters involved ST-131 (with 8/14 clusters caused by a MDR clone) and ST-73 (with 9/15 clusters caused by a non-MDR clone) (Fig. 4 B).
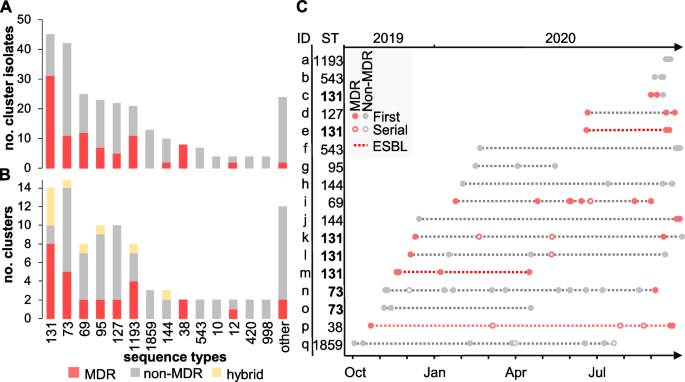
Potential clusters of transmission were defined as groups of isolates from ≥2 patients with ≤17 SNP differences. A Stacked histograms showing the number of MDR and non-MDR cluster isolates according to their ST and B the number of distinct outbreak clusters per ST. Clusters grouping either MDR or non-MDR isolates are shown in red and gray, respectively. Hybrid clusters (i.e . , grouping both MDR and non-MDR isolates) are shown in yellow. STs associated with a single cluster were grouped into others and represent ST-244, ST-394, ST-62, ST-372, ST-404, ST-421, ST-428, ST-538, ST-607, ST-1431, ST-1597, and ST-7887. C Analysis of outbreak clusters (identified on the y -axis with the corresponding ST) involving ≥3 patients ( n = 24) and ordered temporally. The legend describes novel patients (filled circles), serial isolates (open circles), MDR isolates (red-filled circles), non-MDR (black-filled circles), and outbreaks consisting of all ESBL- E. coli isolates (dashed red line)
While the majority of clusters (78 out of 95) were composed of only two patients (an amount of transmission that routine surveillance cannot influence), the remaining outbreaks involved 3 to 12 patients (Fig. 4 C). Temporally, these clusters extended up to 11 months, and lineage ST-131 was once again the most represented, with 5 distinct outbreak clones including two (clusters e and m) that were ESBL-producers (Fig. 4 C). The largest predicted outbreak involved 12 patients (cluster n) and was caused by a ST-73 clone that was largely non-MDR and cultured primarily from urine (Fig. 4 C). The only exception was MDR isolate 836616 which was distinct by only 12 SNPs from non-MDR isolate 822264 from another patient in this cluster and uniquely acquired resistance genes bla TEM-1 , sul2 , aph(3)-lb , and aph(6)-ld (Additional file 3 : Table S3).
Convergence of resistance and virulence determinants in ST-131 E. coli
Considering the role played by ST-131 in both outbreak and sporadic infections, a detailed genetic analysis of the resistance and virulence genes found in these US isolates was performed. A maximum-likelihood core SNP-based phylogeny of all E. coli ST-131 genomes ( n =275) in our dataset resulted in the 3 dominant ST-131 clades: clade A ( n = 59, 21%), clade B ( n = 29, 11%), and clade C ( n = 181, 66%) (Fig. 5 ). Ninety-three percent of clade A isolates carried fimH 41, 84% of clade B carried fimH 22, all subclade B0 isolates carried fimH 27, and 95% of clade C isolates carried the fimH 30 variant. Of note, 19 isolates had non-typeable fimH alleles or a divergent allele designation (Additional file 1 : Table S1).
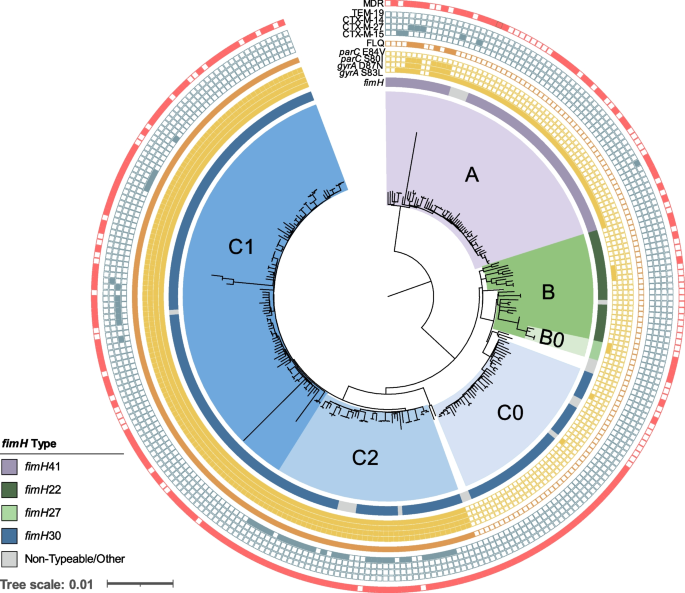
Core genome SNP-based phylogeny of all ST-131 E. coli isolates in our dataset ( n =275). Labels for clades A, B, B0, C0, C2, and C1 are indicated and are colored purple, green, light green, light blue, blue, and dark blue, respectively. Metadata are represented as rings from inner to outer: variations in the fimH gene, presence of point mutations in gyrA and parC (filled yellow square), fluoroquinolone (ciprofloxacin and/or levofloxacin) non-susceptibility (closed orange square), presence of ESBL gene (closed blue square), and multidrug-resistant isolate (red closed square). Non-typeable fimH alleles due to truncation or missing gene were grouped with other rare variants identified (Additional file 1 : Table S1)
Th predominant clade C isolates were further classified into subclades C0 ( n = 38), C1 ( n = 101), and C2 ( n = 42) (Fig. 5 ). Unlike clade A, B, and C0 isolates, which were largely (94%) fluoroquinolone susceptible, 100% of clades C1 and C2 isolates carried double gyrA and parC mutations associated with high-level resistance (Fig. 5 ) [ 56 ]. Furthermore, clade C2 was enriched for ESBL-producing isolates (69% of C2 isolates were ESBL compared to only 9% in other clades) and all carried bla CTX-M-15 . Interestingly, 74% of clade C0 isolates were characterized as MDR (Fig. 5 ) despite being susceptible to fluoroquinolone and cephalosporin antibiotics. This was largely due to a higher prevalence of resistance to aminoglycosides (68% vs. 30% in all), folate pathway inhibitors (68% vs. 38%), and tetracycline (61% vs. 35%) in comparison to other clades (Additional file 2 : Table S2).
In addition to the enrichment of resistance genes, isolates in lineage ST-131 frequently (>80% and p < 0.01) carried virulence-associated genes previously identified and associated with ExPEC E. coli [ 57 , 58 ], including the aerobactin locus ( iucC and iutA otherwise found in ~34% of all E. coli ), a secreted autotransporter toxin ( sat , 25% in the whole population), and an IrgA-like adhesin ( iha , 26% in other E. coli ) (Additional file 4 : Table S4).
Accumulation of virulence genes in non-MDR ST-73 lineage
Together with ST-131, isolates belonging to ST-73 played a prominent role in both sporadic infections and possible cases of nosocomial transmission. However, unlike ST-131, no enrichment of antimicrobial resistance determinants was observed within this lineage, and ST-73 isolates remained largely susceptible to aminoglycosides, cephalosporins, and fluoroquinolones (Fig. 2 , Additional file 2 : Table S2). In contrast, ST-73 isolates contained significantly ( p < 0.01) more virulence-associated genes (between 239 and 314) than ST-131 E. coli (between 201 and 309) [ 13 ] (Additional file 4 : Table S4). Specifically, ST-73 isolates were enriched (>70% vs. <25% in the population as a whole) in uropathogenicity-associated virulence factors involved in invasion and colonization ( pic , hek ), cell lysis ( hlyA ), and adhesion and penetration ( foc/sfa and cnf ) [ 13 , 57 , 59 ] (Additional file 4 : Table S4). When compared to the ST-131 population, ST-73 isolates were significantly enriched in a distinct set of virulence genes most likely contributing to the epidemiological success of the lineage (Additional file 4 : Table S4).
A significant strength of this study lies in the >99% collection of all E. coli isolates from clinical samples over a recent (2019–2020) 12-month period in a network of US military healthcare facilities. Together with comprehensive AST and WGS, this dataset offered a unique opportunity to describe (i) the continued success and emergence of high-risk ExPEC and UPEC lineages, (ii) the regional prevalence of phenotypic resistances (and associated, acquired antibiotic resistance determinants), and (iii) the respective burden of ESBL, MDR, and non-MDR clones in infections of likely nosocomial origin.
Unlike the UK [ 60 , 61 ], Canada [ 18 ], and other regions of the world [ 62 , 63 ], recent genomic surveillance data on circulating E. coli lineages and resistances in the USA is limited. At a global scale, our analysis of this set of US isolates is consistent with previous epidemiological studies demonstrating the predominance (>50% of cases) of ST-69, 73, 95, 127, and 131 pandemic ExPEC lineages [ 16 , 17 ]. E. coli is the world’s leading cause of UTIs, and this is reflected in our collection, where 93% of isolates were from urine samples. The distribution of major lineages observed globally and here also mirrors genomic epidemiology studies of community-acquired (CA)-UTIs across Canada (2012–2015) and from UPEC isolates collected at a Northern California university in 1999–2000 and again in 2016–2017 [ 18 , 19 ]. However, in contrast to these studies, our collection revealed the emergence of ST-1193 fluoroquinolone-resistant E. coli as one of the most prevalent lineages currently circulating in this region of the USA.
Over the past 20 years in the USA, fluoroquinolones have replaced trimethoprim-sulfamethoxazole as the treatment of choice for uncomplicated UTIs [ 64 ]. In our collection, fluoroquinolone non-susceptible isolates largely belonged to only two lineages, ST-131 and ST-1193 (72% between both lineages). For ST-131, numerous studies have described the rapid, global emergence and dominance of subclones with acquired fluoroquinolone resistance mutations (subclade C1/H30-R) and a high prevalence of ESBL enzymes (C2/H30-Rx) [ 16 , 17 ]. In this study, we show that the prevalence of C1 and C2 in the USA (both as an aggregate [52% of ST-131 isolates] and separately with 37% and 15%, respectively) is comparable to estimates from a recent report of a longitudinal collection of E. coli (albeit of bloodstream isolates) from the last two decades in Norway [ 62 ]. Interestingly, these are also similar to earlier US estimates [collection of 261 isolates from 2010 to 2012 [ 16 ]] suggesting the ST-131 population structure has remained relatively stable over the last decade and the overall prevalence of this lineage appears to have plateaued. In contrast, lineage ST-1193, which is the only other known clone driving the spread of fluoroquinolone-resistant E. coli globally [ 65 , 66 , 67 , 68 ], appears to be surging. For example, though the first worldwide cases of ST-1193 only appeared in 2011 [ 68 , 69 ], a recent US-based multicenter surveillance study of 6349 clinical E. coli showed that the fraction of fluoroquinolone-resistant ST-1193 increased from 18 to 25% between 2016 and 2017 [ 67 ]. In our study of isolates from 2019 to 2020, the fraction was 31%, suggesting the rapid rise of ST-1193 is still ongoing. At the molecular level, all ST-1193 in this collection carried three characteristic, non-synonymous mutations resulting in high-level fluoroquinolone resistance; ParC (S80I) and GyrA (D87N and S83L) acquired via homologous recombination from a single transfer event at the origins of that lineage [ 70 ]. In addition, a fourth substitution in ParE (L416F) previously described in ST-1193 lineage was found in all isolates [ 65 ].
In our collection, 5% of primary isolates were ESBL-producers and, of particular concern for treatment regiments, a subset of 74% were co-resistant to the fluoroquinolones. These rates were comparable to the prevalence of resistances observed in a large (>1.5 million isolates), multicenter study of community-onset UTI in the USA over the last decade (6.4% ESBL-producers and 21% fluoroquinolone non-susceptible) [ 71 ]. In contrast, another nationwide US study focused on HAIs during a similar timeframe reported substantially higher rates of resistance to fluoroquinolone (35%) and extended-spectrum cephalosporins (17%) [ 72 ]. Globally, the rate of ESBL -E. coli varies considerably from >40% in regions such as South America, Southeast Asia, India, and China to ~5 to 20% in Europe, Australia, Canada, and the USA [ 73 ]. Furthermore, prevalent lineages carrying ESBLs also vary globally (i.e., ST-648 and ST-410 are underrepresented in our study yet are the most prevalent lineages circulating in intensive care units in Vietnam [ 74 ]). Importantly, ST-1193 was the third most frequent source of ESBL-producers in our collection, with 8% ( n = 7) carrying one of the variously represented alleles ( bla CTX-M-15 , bla CTX-M-27 , bla CTX-M-55 , and first report of bla CTX-M-64 carriage), suggesting multiple introductions. In contrast, ESBL-producers composed 69% of isolates within subclade C2 of ST-131 lineage and all carried the same bla CTX-M-15 , most likely harbored on an IncF-type plasmid as previously characterized [ 75 ]. Finally, while other countries including France [ 76 ], Japan [ 77 ], and Germany [ 78 ] have seen an increase in the recently defined subclade C1-M27 ST-131 [ 77 ] clinical isolates carrying bla CTX-M-27 , we see a low prevalence of subclade C1 bla CTX-M-27 carrying isolates in this study.
While surveillance and infection control efforts are often and understandably (i.e., increased morbidity, mortality, and financial costs) focused on ESBL and MDR E.coli lineages, the global burden of colonization/infection with non-MDR strains (e.g., global lineages ST-73, ST-95, and ST-127) remains invariably higher [ 5 , 62 ]. In fact, in this cohort of 1776 patients, 3 out of 4 individuals were diagnosed with a non-MDR isolate (representing a diversity of E. coli lineages, most of which have yet to be explored). When focusing on patients where in-depth comparative genomics suggested nosocomial origin was likely ( n = 227), a slight increase in the fraction of MDR cases is observed, but the majority (2 out of 3 patients) were still due to a non-MDR clone. In fact, ExPEC pandemic lineage ST-73 was largely comprised of non-MDR isolates and was both one of the most frequent sources of potential clusters of transmission and responsible for the largest predicted outbreak involving 12 patients. While the possibility of transmission happening outside the hospital (e.g., shared long-term facilities or elderly care home) cannot be excluded, these findings highlight the importance of surveilling E. coli isolates with diverse susceptibility profiles as investigations that focus on MDR only are likely to underestimate ongoing outbreaks in the patient population.
To our knowledge, just a single study has performed similar genome-based detection (albeit using a different methodology) of nosocomial transmission on a complete collection of clinical E. coli isolates [ 9 ]. That study examined stool samples from 97 inpatients over a 6-month period at a single UK hospital. Similar to our findings, the two largest clusters identified spanned the entirety of the study period and were caused by the nosocomial spread of non-MDR isolates. Interestingly, these clones were identified as ST-7095 (7 patients, 29 isolates) and ST-635 (4 patients, 18 isolates) [ 9 ], two lineages comprised within phylogroup A that were not detected in our sampling. Whether the epidemic success of these non-MDR lineages simply stems from their overall abundance or could result from the acquisition of virulence/colonization factors (as observed here for ST-73) remains to be fully characterized.
Conclusions
By capturing all clinical isolates for a full year, this study provides a rare and contemporary survey of the genomic landscape of MDR and non-MDR E. coli lineages in a large healthcare network in the Northeast US. While pandemic ST-131 and expanding ST-1193 lineages (both characterized by high rates of co-resistance to fluoroquinolones and extended-spectrum cephalosporins) warrant particular surveillance, our findings also indicate that non-MDR lineages play a significant role in nosocomial transmission. With WGS developing as a near-routine technology in infection control, such improved understanding of the epidemiology of hospital-acquired pathogens is critical for maximum effectiveness at reducing infections and healthcare-associated costs.
Availability of data and materials
Both genomic assemblies and raw sequencing data of all isolates analyzed in this study are publicly available in the NCBI database under the BioProject number PRJNA809394 [ 79 ].
Abbreviations
Extended-spectrum β-lactamase
Multidrug resistant
Multilocus sequence typing
Sequence type
Core genome multilocus sequence typing
National Center for Biotechnology Information
Vihta KD, Stoesser N, Llewelyn MJ, Quan TP, Davies T, Fawcett NJ, et al. Trends over time in Escherichia coli bloodstream infections, urinary tract infections, and antibiotic susceptibilities in Oxfordshire, UK, 1998–2016: a study of electronic health records. Lancet Infect Dis. 2018;18(10):1138–49.
Article Google Scholar
Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269–84.
Article CAS Google Scholar
Critchley IA, Cotroneo N, Pucci MJ, Mendes R. The burden of antimicrobial resistance among urinary tract isolates of Escherichia coli in the United States in 2017. PLoS One. 2019;14(12):e0220265.
Zhen X, Lundborg CS, Sun X, Hu X, Dong H. Economic burden of antibiotic resistance in ESKAPE organisms: a systematic review. Antimicrob Resist Infect Control. 2019;8:137.
Naylor NR, Pouwels KB, Hope R, Green N, Henderson KL, Knight GM, et al. The health and cost burden of antibiotic resistant and susceptible Escherichia coli bacteraemia in the English hospital setting: a national retrospective cohort study. PLoS One. 2019;14(9):e0221944.
Sader HS, Castanheira M, Flamm RK, Jones RN. Antimicrobial activities of ceftazidime-avibactam and comparator agents against Gram-negative organisms isolated from patients with urinary tract infections in U.S. medical centers, 2012 to 2014. Antimicrob Agents Chemother. 2016;60(7):4355–60.
Karlowsky JA, Kelly LJ, Thornsberry C, Jones ME, Sahm DF. Trends in antimicrobial resistance among urinary tract infection isolates of Escherichia coli from female outpatients in the United States. Antimicrob Agents Chemother. 2002;46(8):2540–5.
Sawatwong P, Sapchookul P, Whistler T, Gregory CJ, Sangwichian O, Makprasert S, et al. High burden of extended-spectrum β-lactamase–producing Escherichia coli and Klebsiella pneumoniae bacteremia in older adults: a seven-year study in two rural Thai provinces. Am J Trop Med Hyg. 2019;100(4):943–51.
Ludden C, Coll F, Gouliouris T, Restif O, Blane B, Blackwell GA, et al. Defining nosocomial transmission of Escherichia coli and antimicrobial resistance genes: a genomic surveillance study. Lancet. Microbe. 2021;0(0). 2021;2(9):e472-80.
Raphael E, Glymour MM, Chambers HF. Trends in prevalence of extended-spectrum beta-lactamase-producing Escherichia coli isolated from patients with community- and healthcare-associated bacteriuria: results from 2014 to 2020 in an urban safety-net healthcare system. Antimicrob Resist Infect Control. 2021;10(1):118.
Centers for Disease Control and Prevention (U.S.). Antibiotic resistance threats in the United States, 2019 [Internet]. Centers for Disease Control and Prevention (U.S.); 2019. Available from: https://stacks.cdc.gov/view/cdc/82532 . [Cited 2021 May 24].
Denamur E, Clermont O, Bonacorsi S, Gordon D. The population genetics of pathogenic Escherichia coli. Nat Rev Microbiol. 2021;19(1):37-45.
Biggel M, Xavier BB, Johnson JR, Nielsen KL, Frimodt-Møller N, Matheeussen V, et al. Horizontally acquired papGII-containing pathogenicity islands underlie the emergence of invasive uropathogenic Escherichia coli lineages. Nat Commun. 2020;11(1):5968.
Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2(2):123–40.
Pitout JDD, DeVinney R. Escherichia coli ST131: a multidrug-resistant clone primed for global domination. F1000Research. 2017;6:F1000 Faculty Rev-195.
Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, et al. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio. 2013;4(6): e00377-13.
Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis. 2013;207(6):919–28.
Fibke CD, Croxen MA, Geum HM, Glass M, Wong E, Avery BP, et al. Genomic epidemiology of major extraintestinal pathogenic Escherichia coli lineages causing urinary tract infections in young women across Canada. Open Forum. Infect Dis Ther. 2019;6(11):ofz431.
Google Scholar
Yamaji R, Rubin J, Thys E, Friedman CR, Riley LW. Persistent pandemic lineages of uropathogenic Escherichia coli in a college community from 1999 to 2017. Diekema DJ, editor. J Clin Microbiol. 2018;56(4):e01834–17.
Kallonen T, Brodrick HJ, Harris SR, Corander J, Brown NM, Martin V, et al. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017;27(8):1437–49.
Duval A, Obadia T, Boëlle PY, Fleury E, Herrmann JL, Guillemot D, et al. Close proximity interactions support transmission of ESBL-K. pneumoniae but not ESBL-E. coli in healthcare settings. PLoS Comput Biol. 2019;15(5):e1006496.
CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 11th ed. CLSI standard M07. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
Galac MR, Snesrud E, Lebreton F, Stam J, Julius M, Ong AC, et al. A diverse panel of clinical Acinetobacter baumannii for research and development. Antimicrob Agents Chemother. 2020;64(10):e00840–20 /aac/64/10/AAC.00840-20.atom.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9.
Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257.
Waters NR, Abram F, Brennan F, Holmes A, Pritchard L. Easy phylotyping of Escherichia coli via the EzClermont web app and command-line tool. Access Microbiol. 2020;2(9):acmi000143.
Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5(1):58–65.
Seeman T. mlst - scan contig files against traditional PubMLST typing schemes. Available from: https://github.com/tseemann/mlst . [Accessed 2020 Jan 4].
Jolley KA, Bray J, Maiden MC. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;(3):124 https://pubmlst.org/ . Accessed 4 January 2020.
Zhou Z, Alikhan NF, Mohamed K, Fan Y, the Agama Study Group, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30(1):138–52.
Quijada NM, Rodríguez-Lázaro D, Eiros JM, Hernández M. TORMES: an automated pipeline for whole bacterial genome analysis. Bioinformatics. 2019;35(21):4207–12.
Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol. 2015;53(8):2410–26.
Roer L, Tchesnokova V, Allesøe R, Muradova M, Chattopadhyay S, Ahrenfeldt J, et al. Development of a web tool for Escherichia coli subtyping based on fimH alleles. J Clin Microbiol. 2017;55(8):2538–43.
Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63(11):e00483–19.
Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, Keane JA, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genomics. 2017;3(10):e000131.
Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–903.
Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44(D1):D694–7.
Biggel, M. EcVGDB. Zenodo [Internet]. 2020; Available from: https://doi.org/10.5281/zenodo.4079473 .
Seemann T. ABRicate - mass screening of contigs for antimicrobial resistance or virulence genes. Available from: https://github.com/tseemann/abricate . [Accessed 2022 Feb 23].
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3.
Seeman T. snippy - rapid haploid variant calling and core genome alignment. Available from: https://github.com/tseemann/abricate . [Accessed 2019 Sep 25].
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963.
Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3.
Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–9.
Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio. 2016;7(2):e02162.
Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, et al. Updating benchtop sequencing performance comparison. Nat Biotechnol. 2013;31(4):294–6.
Muloi DM, Wee BA, McClean DMH, Ward MJ, Pankhurst L, Phan H, et al. Population genomics of Escherichia coli in livestock-keeping households across a rapidly developing urban landscape. Nat Microbiol. 2022;7(4):581–9.
Johnson JR, Johnston BD, Porter SB, Clabots C, Bender TL, Thuras P, et al. Rapid emergence, subsidence, and molecular detection of Escherichia coli sequence type 1193- fimH64 , a new disseminated multidrug-resistant commensal and extraintestinal pathogen. J Clin Microbiol. 2019;57(5):e01664–18.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Mshana SE, Falgenhauer L, Mirambo MM, Mushi MF, Moremi N, Julius R, et al. Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect Dis. 2016;16(1):187.
Day MJ, Hopkins KL, Wareham DW, Toleman MA, Elviss N, Randall L, et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect Dis. 2019;19(12):1325–35.
Guenther S, Semmler T, Stubbe A, Stubbe M, Wieler LH, Schaufler K. Chromosomally encoded ESBL genes in Escherichia coli of ST38 from Mongolian wild birds. J Antimicrob Chemother. 2017;72(5):1310–3.
Kondratyeva K, Salmon-Divon M, Navon-Venezia S. Meta-analysis of pandemic Escherichia coli ST131 plasmidome proves restricted plasmid-clade associations. Sci Rep. 2020;10(1):36.
Xia L, Liu Y, Xia S, Kudinha T, Xiao SN, Zhong NS, et al. Prevalence of ST1193 clone and IncI1/ST16 plasmid in E-coli isolates carrying blaCTX-M-55 gene from urinary tract infections patients in China. Sci Rep. 2017;7(1):44866.
Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51(5):1109–17.
Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, Frej-Madrzak M, Ksiazczyk M, Bugla-Ploskonska G, et al. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 2019;11(1):10.
Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, et al. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio. 2016;7(2):e00347–16.
Jahandeh N, Ranjbar R, Behzadi P, Behzadi E. Uropathogenic Escherichia coli virulence genes: invaluable approaches for designing DNA microarray probes. Cent Eur. J Urol. 2015;68(4):452-458.
Brodrick HJ, Raven KE, Kallonen T, Jamrozy D, Blane B, Brown NM, et al. Longitudinal genomic surveillance of multidrug-resistant Escherichia coli carriage in a long-term care facility in the United Kingdom. Genome Med. 2017;9(1):70.
Lipworth S, Vihta KD, Chau KK, Kavanagh J, Davies T, George S, et al. Ten years of population-level genomic Escherichia coli and Klebsiella pneumoniae serotype surveillance informs vaccine development for invasive infections. Clin Infect Dis. 2021;73(12):2276–82.
Gladstone RA, McNally A, Pöntinen AK, Tonkin-Hill G, Lees JA, Skytén K, et al. Emergence and dissemination of antimicrobial resistance in Escherichia coli causing bloodstream infections in Norway in 2002–17: a nationwide, longitudinal, microbial population genomic study. Lancet Microbe. 2021;2(7):e331–41.
Mahazu S, Sato W, Ayibieke A, Prah I, Hayashi T, Suzuki T, et al. Insights and genetic features of extended-spectrum beta-lactamase producing Escherichia coli isolates from two hospitals in Ghana. Sci Rep. 2022;12(1):1843.
Kallen AJ, Welch HG, Sirovich BE. Current antibiotic therapy for isolated urinary tract infections in women. Arch Intern Med. 2006;166(6):635.
Johnson TJ, Elnekave E, Miller EA, Munoz-Aguayo J, Flores Figueroa C, Johnston B, et al. Phylogenomic analysis of extraintestinal pathogenic Escherichia coli sequence type 1193, an emerging multidrug-resistant clonal group. Antimicrob Agents Chemother. 2019;63(1):e01913–8.
Valenza G, Werner M, Eisenberger D, Nickel S, Lehner-Reindl V, Höller C, et al. First report of the new emerging global clone ST1193 among clinical isolates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli from Germany. J Glob Antimicrob Resist. 2019;17:305–8.
Tchesnokova VL, Rechkina E, Larson L, Ferrier K, Weaver JL, Schroeder DW, et al. Rapid and extensive expansion in the United States of a new multidrug-resistant Escherichia coli clonal group, sequence type 1193. Clin Infect Dis. 2019;68(2):334–7.
Wu J, Lan F, Lu Y, He Q, Li B. Molecular characteristics of ST1193 clone among phylogenetic group B2 non-ST131 fluoroquinolone-resistant Escherichia coli. Front Microbiol. 2017;8:2294.
Nguyen Q, Nguyen TTN, Pham P, Chau V, Nguyen LPH, Nguyen TD, et al. Genomic insights into the circulation of pandemic fluoroquinolone-resistant extra-intestinal pathogenic Escherichia coli ST1193 in Vietnam. Microb. Genomics. 2021;7(12):000733.
Tchesnokova V, Radey M, Chattopadhyay S, Larson L, Weaver JL, Kisiela D, et al. Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene loci. Proc Natl Acad Sci. 2019;116(29):14740–8.
Kaye KS, Gupta V, Mulgirigama A, Joshi AV, Scangarella-Oman NE, Yu K, et al. Antimicrobial resistance trends in urine Escherichia coli isolates from adult and adolescent females in the United States from 2011 to 2019: rising ESBL strains and impact on patient management. Clin Infect Dis. 2021;73(11):1992–9.
Kourtis AP, Sheriff EA, Weiner-Lastinger LM, Elmore K, Preston LE, Dudeck M, et al. Antibiotic multidrug resistance of Escherichia coli causing device- and procedure-related infections in the United States reported to the National Healthcare Safety Network, 2013–2017. Clin Infect Dis. 2021;73(11):e4552-59.
Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–55.
Roberts LW, Hoi LT, Khokhar FA, Hoa NT, Giang TV, Bui C, et al. Genomic characterisation of multidrug-resistant Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii in two intensive care units in Hanoi, Viet Nam: a prospective observational cohort study. Lancet Microbe. 2022;3(11):e857-e866.
Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72(8):2145–55.
Birgy A, Bidet P, Levy C, Sobral E, Cohen R, Bonacorsi S. CTX-M-27-producing Escherichia coli of sequence type 131 and clade C1-M27, France. Emerg. Infect Dis. 2017; 23(5):885.
Matsumura Y, Pitout JDD, Gomi R, Matsuda T, Noguchi T, Yamamoto M, et al. Global Escherichia coli sequence type 131 clade with bla CTX-M-27 gene. Emerg Infect Dis. 2016;22(11):1900–7.
Ghosh H, Doijad S, Falgenhauer L, Fritzenwanker M, Imirzalioglu C, Chakraborty T. bla CTX-M-27 –encoding Escherichia coli sequence type 131 lineage C1-M27 clone in clinical isolates. Germany Emerg Infect Dis. 2017;23(10):1754–6.
Mills EG, Martin MJ, et al. A one-year genomic investigation of Escherichia coli epidemiology and nosocomial spread at a large U.S. healthcare network. Natl Cent Biotechnol Inf. 2022; Available from: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA809394 .
Download references
Acknowledgements
The authors are thankful to all the staff of the MRSN and the clinical microbiology laboratory of the WRNMMC. The manuscript has been reviewed by the Walter Reed Army Institute of Research and there is no objection to its presentation. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or reflecting the views of the Department of the Army or the Department of Defense.
This study was funded by the Armed Forces Health Surveillance Division (AFHSD), Global Emerging Infections Surveillance (GEIS) Branch ProMIS ID P001_20_WR (to P. Mc Gann).
Author information
Emma G. Mills and Melissa J. Martin contributed equally to this work.
Authors and Affiliations
Multidrug-Resistant Organism Repository and Surveillance Network, Walter Reed Army Institute of Research, Silver Spring, MD, USA
Emma G. Mills, Melissa J. Martin, Ting L. Luo, Ana C. Ong, Rosslyn Maybank, Brendan W. Corey, Casey Harless, Lan N. Preston, Joshua A. Rosado-Mendez, Yoon I. Kwak, Jason W. Bennett, Patrick T. Mc Gann & Francois Lebreton
Department of Pathology, Walter Reed National Military Medical Center, Bethesda, MD, USA
Scott B. Preston & Michael G. Backlund
You can also search for this author in PubMed Google Scholar
Contributions
P.T.M. designed the research; P.T.M. E.G.M., M.J.M., T.L.L., R.M., B.W.C., C.H., A.C.O, L.N.P, J.A.R-M, G.G.B, S.B.P., and Y.I.K performed the research. E.G.M., M.J.M., J.W.B, P.T.M., and F.L. analyzed the research; E.G.M., M.J.M., and F.L. wrote the paper with input from all authors. All authors read and approved the final version of the manuscript.
Corresponding authors
Correspondence to Patrick T. Mc Gann or Francois Lebreton .
Ethics declarations
Ethics approval and consent to participate.
The isolates and clinical information were collected as part of the public health surveillance activities of the MRSN, as determined by the WRAIR branch director and Human Subjects Protection Branch (HSPB) which granted ethical approval. Informed patient consent was waived as samples were taken under a hospital surveillance framework for routine sampling. The research conformed to the principles of the Helsinki Declaration.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: table s1..
Basic Isolate Metadata.
Additional file 2: Table S2.
Antibiotic Susceptibility Testing.
Additional file 3: Table S3.
AMR Genetic Characteristics.
Additional file 4: Table S4.
Virulence Associated Genes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Mills, E.G., Martin, M.J., Luo, T.L. et al. A one-year genomic investigation of Escherichia coli epidemiology and nosocomial spread at a large US healthcare network. Genome Med 14 , 147 (2022). https://doi.org/10.1186/s13073-022-01150-7
Download citation
Received : 15 June 2022
Accepted : 13 December 2022
Published : 30 December 2022
DOI : https://doi.org/10.1186/s13073-022-01150-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Escherichia coli
- Genomic epidemiology
- Antibiotic resistance
Genome Medicine
ISSN: 1756-994X
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Section 2: Fundamentals
Case study: e. coli, what would you do.
You’re the state epidemiologist for infectious diseases. More than forty children in county X contracted what appears to be E. coli 0157. When the state health department announces to the press that there is an E. coli outbreak and that the source is a famous restaurant chain, media interest skyrockets. The public information officer for the state health department asks you to continue fielding questions from reporters, but lab tests aren’t telling you anything new.
What They Did
In January 1993, the Washington State Health Department investigated a suspected E. coli 0157 H7, which made more than forty children in the Puget Sound region ill. Initially, the press paid little attention. But when the department announced to the press that it was dealing with an E. coli outbreak—and that the source of the outbreak was the restaurant chain Jack in the Box—the media interest skyrocketed. This case involved a potentially fatal disease, children, and a national restaurant chain. Suddenly, the department was dealing with a national news story.
John Kobayashi, then the State Epidemiologist for Infectious Diseases, found that his time was consumed with responding to the press during the week following the announcement. In this video, Dr. Kobayashi describes his experience with crisis emergency risk communication.
At the end of the week I was pretty tired because we made the public announcement, as I recall it was on the beginning of the week, about Jack in the Box being related to the outbreak, and that the food was being quarantined, and so on, and so on. It was really, really big news at that time. And so we were very, very busy that week investigating the outbreak and also responding to media questions.
So I was ready to have a rest at the end of that week. But my public affairs officer, Dean Owen, talked to me and he said, “Don’t stop talking to the media. And it’s really important to continue your message.” And I didn’t really understand that because I thought we had said everything we were going to say. And we were waiting for culture results and the data to be finalized and so on.
And he said, “No, you need to keep talking to the media. This is a very big issue. It’s a national story and people need more information, even if it’s the same information.” And he said that if I stop talking to the media, then the media would be looking for other people to talk to and that information might not be as up-to-date and accurate as it ought to be.
And as chance would have happened, we were dealing with another problem by that weekend. There were about 60 children who had the infection and these children had been in day care centers. And E. coli 0157 can be transmitted in many ways, one of which is through contaminated food, but also it can be passed from person to person very easily, especially in situations like day care centers. So that became a big concern of ours.
So that weekend I had about four interviews with the media talking about the importance of hand washing all of the time, but especially when you're ill with something, especially when your ill with something like E. coli 0157. So I talked about hand washing, hand washing, hand washing during that weekend to the media. And that was actually a very good thing. There were three children who died of E. coli 0157 in that outbreak. And actually, two of the children who died were not direct consumers of the hamburgers. They were contacts of people who had consumed the hamburgers. So secondary transmission was very important.
So that message was carried through the weekend. And although “secondary transmission” is more an epidemiological term, it became a household word in Washington state. And everybody knew what it was.
And I am convinced to this day that we probably reduced the number of secondary cases because of the messaging we did that weekend.
- NEXT: Section Summary
- Open access
- Published: 06 July 2024
Impact of the COVID-19 pandemic on extended-spectrum β-lactamase producing Escherichia coli in urinary tract and blood stream infections: results from a nationwide surveillance network, Finland, 2018 to 2022
- Heikki Ilmavirta 1 , 2 , 3 ,
- Jukka Ollgren 3 ,
- Kati Räisänen 3 ,
- Tuure Kinnunen 1 , 2 ,
- Antti Juhani Hakanen 4 ,
- Kaisu Rantakokko-Jalava 4 ,
- Jari Jalava 3 &
- Outi Lyytikäinen 3
Antimicrobial Resistance & Infection Control volume 13 , Article number: 72 ( 2024 ) Cite this article
144 Accesses
Metrics details
Before the COVID-19 pandemic there has been a constant increase in antimicrobial resistance (AMR) of Escherichia coli , the most common cause of urinary tract infections and bloodstream infections. The aim of this study was to investigate the impact of the COVID-19 pandemic on extended-spectrum β-lactamase (ESBL) production in urine and blood E. coli isolates in Finland to improve our understanding on the source attribution of this major multidrug-resistant pathogen.
Susceptibility test results of 564,233 urine (88.3% from females) and 23,860 blood E. coli isolates (58.8% from females) were obtained from the nationwide surveillance database of Finnish clinical microbiology laboratories. Susceptibility testing was performed according to EUCAST guidelines. We compared ESBL-producing E. coli proportions and incidence before (2018–2019), during (2020–2021), and after (2022) the pandemic and stratified these by age groups and sex.
The annual number of urine E. coli isolates tested for antimicrobial susceptibility decreased 23.3% during 2018–2022 whereas the number of blood E. coli isolates increased 1.1%. The annual proportion of ESBL-producing E. coli in urine E. coli isolates decreased 28.7% among males, from 6.9% (average during 2018–2019) to 4.9% in 2022, and 28.7% among females, from 3.0 to 2.1%. In blood E. coli isolates, the proportion decreased 32.9% among males, from 9.3 to 6.2%, and 26.6% among females, from 6.2 to 4.6%. A significant decreasing trend was also observed in most age groups, but risk remained highest among persons aged ≥ 60 years.
Conclusions
The reduction in the proportions of ESBL-producing E. coli was comprehensive, covering both specimen types, both sexes, and all age groups, showing that the continuously increasing trends could be reversed. Decrease in international travel and antimicrobial use were likely behind this reduction, suggesting that informing travellers about the risk of multidrug-resistant bacteria, hygiene measures, and appropriate antimicrobial use is crucial in prevention. Evaluation of infection control measures in healthcare settings could be beneficial, especially in long-term care.
Antimicrobial resistance (AMR) has emerged as one of the leading public health threats in the 21st century [ 1 , 2 ]. AMR is accelerated by misuse or overuse of antimicrobials and poor infection prevention and control (IPC) [ 3 ]. Hence, antimicrobial stewardship programs and IPC have been used as mitigation strategies against AMR. In addition, several other factors may contribute to AMR, such as the presence of multidrug-resistant (MDR) bacteria in livestock and agricultural products, and increasing foreign travel [ 4 ], particularly to countries with a high prevalence of MDR bacteria. The onset of the COVID-19 pandemic affected healthcare systems, causing major disruptions that threaten the effectiveness of IPC and antimicrobial stewardship strategies [ 5 , 6 ]. The COVID-19 pandemic also complicated AMR surveillance and research, as changes in healthcare delivery, improved IPC measures related to the pandemic, and reduced national and international travel may have reduced the selection of pathogens resistant to antimicrobials in a short term [ 7 ]. However, opposite impacts could also be seen if antimicrobials have been used more frequently and inappropriately during the pandemic.
Escherichia coli is the leading cause of urinary tract infections (UTI) and bloodstream infections (BSI) worldwide, causing substantial and increasing burden of disease, especially among elderly people [ 8 , 9 , 10 , 11 , 12 ]. The emergence of AMR among E. coli causes major concern, as infections caused by MDR E. coli are more challenging to treat, conferring a higher risk of bacteraemia and death [ 13 ]. Extended-spectrum β-lactamase (ESBL) production provides resistance to many clinically important antimicrobials, including third-generation cephalosporins (3GC), which are widely used as the first-line empirical treatment in severe E. coli infections, such as pyelonephritis or BSI. Several recent surveillance reports have demonstrated a decrease in the proportion of ESBL-producing or 3GC-resistant E. coli during the pandemic years 2020–2022 [ 14 , 15 , 16 , 17 ]. Also, in the latest report of European Antimicrobial Resistance Surveillance Network (EARS-Net), there was an overall decreasing trend of 3GC-resistance in invasive E. coli isolates [ 18 ]. However, these surveillance reports have rarely covered both urine and blood isolates or evaluated the proportions and risk in different age groups and sex.
Our previous study covering the years preceding the COVID-19 pandemic (2008–2019) demonstrated an average annual increase (AAI) of around 9% in the proportion of ESBL-producing E. coli among urine and blood E. coli isolates in Finland, and this increase was similar in all age groups regardless of sex [ 19 ]. In the current study, we investigated the impact of the COVID-19 pandemic on the epidemiology of ESBL-producing E. coli and analysed the trends in the proportion of ESBL-producing isolates among E. coli isolated from blood or urine cultures in different age groups and both sexes during and after the COVID-19 pandemic. We also assessed the changes in the incidence of ESBL-producing E. coli during the study period.
The national Finres database [ 20 ] contains antimicrobial susceptibility test results of 20 common clinically important bacteria under surveillance in Finland, including E. coli [ 19 ]. For each bacterial species, only the first isolate with a susceptibility test result per sample type and patient is reported to this database annually. Information collected includes bacterial name, susceptibility test results for selected antimicrobials (disc diffusion, minimum inhibitory concentration, interpretation of the test result, and/or confirmed resistance mechanism), age and sex (male or female), and date and type of specimen. Antimicrobial susceptibility tests including phenotypical ESBL screening and confirmation were performed and interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [ 21 ]. The described data are reported annually by all Finnish clinical microbiology laboratories, covering all healthcare districts in Finland. All laboratories are government licenced and participate in international and national external quality assessment programmes including the EARS-Net quality control scheme. During 2018–2022, the annual number of laboratories reporting blood cultures varied between 15 and 19 and the number of those reporting urine cultures between 15 and 21. The Finres database covered 95% (range by year 87–100%) of all blood [ 22 ] and approximately 90% of all urine culture isolates sampled in Finland during the study period.
Analysis and statistics
To minimize bias, we excluded one laboratory accounting annually an average of 9.9% of all blood and 7.8% of all urine isolates in the Finres database during 2018–2021, since the laboratory was not able to report susceptibility test results for 2022 due to technical reasons.
We calculated the annual proportions of ESBL-producing E. coli isolates from all urine and blood E. coli i solates for different sexes and age groups, and the annual proportion of fluoroquinolone resistant ESBL-producing E. coli isolates, defined as ESBL-producing and resistant to moxifloxacin, levofloxacin, ciprofloxacin, and/or norfloxacin. We also calculated the annual incidences of ESBL-producing E. coli per 100,000 inhabitants. To compare observed trends over time and between age groups and sex, we applied a binomial regression model with log link and with or without Newey–West standard errors, which take into account the possible autocorrelation conditional on the chosen trend. For average annual decreases (AAD) and trends, we calculated 95% compatibility intervals (CI) and p values, p values of < 0.05 were considered statistically significant. In addition, we calculated the mean annual proportion of ESBL-producing E. coli isolates during 2018–2019 and compared it to the proportion in 2022 to assess the relative and absolute change during the pandemic. Data were analysed using SPSS Statistics 25 (IBM, .ibm.com) and Stata 17.0 (StataCorp LLC, .stata.com).
During 2018–2022, a total of 848,168 urine culture and 56,788 blood culture bacterial isolates were identified in the Finres database; 93.4% (792,526/848,168) of the urine isolates and 92.0% (52,219/56,788) of the blood isolates were included in our analyses. Of the included isolates, 71.2% (564,233/792,526) of the urine isolates and 45.7% (23,860/52,219) of the blood isolates were identified as E. coli .
The total annual number of all urine isolates decreased by 22.5% during the study period, from 176,904 in 2018 to 137,013 in 2022, but remained stable for blood isolates (9,970 in 2018, 10,947 in 2019, and 10,662 in 2022). Similarly, the total annual number of urine E. coli isolates tested for antimicrobial susceptibility decreased by 23.3%, from 125,315 in 2018 to 96,123 in 2022, whereas the number of blood isolates increased by 1.1%, from 4,523 in 2018 to 4575 in 2022. Of urine E. coli isolates, 88.3% (498,162/564,233) were from females and 11.7% (66,071/564,233) from males, and 58.8% (14,020/23,860) of blood E. coli isolates were from females and 41.2% (9,840/23,860) from males. The proportions of urine and blood E. coli isolates from patients aged ≥ 60 years were 71.1% (401,044/564,233) and 84.5% (20,170/23,860), respectively.
For all E. coli isolates, information of their ESBL status was available. In addition, susceptibility test result for at least one fluoroquinolone was available for 98.8% (557,518/564,233) of the urine and 99.8% (23,809/23,860) of the blood E. coli isolates.
During 2019–2022, a significant decreasing trend in the annual proportion of ESBL-producing E. coli in urine and blood E. coli isolates was observed in both males and females (Fig. 1 and Supplementary Table S1 ). In urine isolates, the decrease averaged 27.6% among both sexes from 2018/19 to 2022: among males from 6.9 to 4.9% (AAD: 12.1%, 95%CI: 9.3–14.7%, p < 0.01) and among females from 3.0 to 2.1% (AAD: 11.7%, 95%CI: 10.1–13.2%, p < 0.01). In blood isolates, the proportion decreased by an average of 29.0% from 2018/19 to 2022: among males from 9.3 to 6.2% (AAD: 11.3%, 95%CI: 4.9–17.2%, p < 0.01) and among females from 6.2 to 4.6% (AAD: 12.0%, 95%CI: 5.6–18.0%, p < 0.01). The AAD values were similar for both urine and blood isolates for both sexes during the study period. Notably, the annual proportion of ESBL-producing E. coli was constantly higher in blood than in urine E. coli isolates and higher in males than in females.

The annual proportion of extended-spectrum β-lactamase-producing Escherichia coli in blood and urine E. coli isolates among males and females, Finland, 2018–2022. AAD: average annual decrease; CI: compatibility interval; ESBL+: extended-spectrum β-lactamase-producing Escherichia coli ; RelD: relative decrease
Importantly, the annual proportion of ESBL-producing E. coli in urine and blood E. coli isolates decreased in all age groups during 2019–2022, except for blood isolates from males aged 0–19 years and females aged 20–39 years (Fig. 2 and Supplementary Table S1 ). In urine isolates, the decreasing trend was statistically significant in all age groups, except for males aged 20–39 years, but in blood isolates significant only in age groups of 60–79 and ≥ 80 years among both sexes. When considering the 95% CIs, the significant trends (AADs) were very similar in different age groups for both sexes.

The annual proportion of extended-spectrum β-lactamase-producing Escherichia coli in urine E. coli isolates among ( A ) males and ( B ) females and in blood E. coli isolates among ( C ) males and ( D ) females, Finland, 2019–2022. AAD: average annual decrease; CI: compatibility interval; ESBL+: extended-spectrum β-lactamase-producing Escherichia coli
Quarterly analysis shows that, the proportion of ESBL-producing E. coli isolates started to decrease during quarter 2 and 3 of 2020 for urine and during quarter 2 of 2020 for blood isolates – immediately after the onset of the pandemic (quarter 1 of 2020) (Fig. 3 A and C). In quarter 3 and 4 of 2022, this decrease stabilized for urine isolates and started to increase again for blood isolates. Although the numbers of urine E. coli isolates tested decreased during the pandemic years, quarterly testing activity remained rather unchanged throughout the study period (Fig. 3 B).

The quarterly analysis of the proportions of extended-spectrum β-lactamase-producing Escherichia coli in ( A ) urine and ( C ) blood E. coli isolates and the number of urine ( B ) and blood ( D ) E. coli isolates tested, Finland, 2018–2022. ESBL+: extended-spectrum β-lactamase-producing Escherichia coli , Q1-Q4: quarter 1–4
The incidence of ESBL-producing E. coli in urine and blood cultures decreased in most age groups during 2019–2022 among both sexes (Fig. 4 and Supplementary Table S2 ). However, the incidence decreased overall more in urine isolates than in blood isolates. In urine cultures, the decrease averaged 44.8% from 2018/19 to 2022: among males from 38.6 per 100.000 inhabitants in 2018/19 to 23.6 in 2022 (AAD: 17.2%, 95%CI: 14.5–19.7%, p < 0.01) and among females from 131.3 to 70.3 (AAD: 20.0%, 95%CI: 18.6–21.4%, p < 0.01). In blood cultures, the incidence decreased by an average of 31.6% from 2018/19 to 2022: among males from 7.0 to 4.9 (AAD: 11.1%, 95%CI: 4.5–17.2%, p < 0.01) and among females from 7.0 to 4.7 (AAD: 14.6%, 95%CI: 8.2–20.6%, p < 0.01). This decreasing trend was significant in all age groups for urine isolates, but for blood isolates only in males and females aged ≥ 60 years. When considering the 95% CIs, the significant trends (AADs) were very similar in different age groups for both sexes. The largest decrease in the incidence was observed in the two oldest age groups (60–79 and ≥ 80 years), being particularly prominent in persons aged ≥ 80 years.

The annual incidence of extended-spectrum β-lactamase-producing Escherichia coli (numbers per 100,000 inhabitants) in urine cultures among ( A ) males and ( B ) females and in blood cultures among ( C ) males and ( D ) females, Finland, 2019–2022. AAD: average annual decrease; CI: compatibility interval; ESBL+: extended-spectrum β-lactamase-producing Escherichia coli
The proportion of fluoroquinolone-resistant isolates among ESBL-producing E. coli decreased only slightly during 2019–2022: in urine isolates from 67.6 to 64.5% and in blood isolates from 73.1 to 66.4%. Of note, E. coli isolates resistant (R) or susceptible with increased exposure (I) to meropenem or imipenem were rare during the study period: a total of 28 isolates in urine (range by year, 2–8; 0.014% (28/200,605)) and 5 isolates in blood (range by year, 0–2; 0.019% (5/25,855)).
Our study based on the national surveillance data indicates that the annual proportions of ESBL-producing isolates among E. coli from urine and blood cultures significantly decreased after the onset of the COVID-19 pandemic during 2019–2022. Concurrently, the incidence of ESBL-producing E. coli significantly decreased in urine cultures in both sexes in all age groups, and also in blood cultures of both males and females ≥ 60 years of age. In addition, we observed a clear decrease in the annual number of urine isolates reported to the surveillance database during the pandemic. However, for blood isolates, there was a slight increase during this timeframe. Furthermore, for ESBL-producing E. coli isolates, coincident resistance to fluoroquinolones remained high during the study period.
Our study shows that, the observed decreasing trends in the proportion of ESBL-producing E. coli were more than a mirror image of the trends observed in our previous study in Finland covering the pre-pandemic years 2008–2019 (AAD during 2019–2022: 11.3% in urine and 11.4% in blood (Supplementary table S2 ) vs. AAI during 2008–2019: 8.9% in urine and 8.7% in blood) [ 19 ]. The lowest proportions observed in this study in 2022 were roughly at the same level as observed in 2015, 5 years before the onset of the pandemic. Notably, the previously observed differences between sexes and sample types in the levels of the proportions remained the same, with the proportion of ESBL-producing isolates being higher among males than females and higher in blood isolates than in urine isolates. Our results are also partly paralleled by three previous studies [ 23 , 24 , 25 ]. In France, an overall significant decrease in ESBL production among E. coli isolates from clinical samples of primary care patients and nursing home residents was reported after the national lockdown on the 11th of May 2020 [ 23 ]. The decrease was statistically significant for urine cultures, females, and the age groups of 5–19, 40–64, and > 60 years. In Ontario, Canada, a decreasing trend for ESBL-producing E. coli in urine cultures from community patients and patients in long-term care facilities (LTCF) during the COVID-19 pandemic was also observed [ 24 ]. However, the study periods in these two studies were shorter than ours. In the Netherlands, in hospitalised patients, a significantly lower prevalence of ESBL-producing E. coli and Klebsiella pneumoniae was observed from June to August 2022 compared to the pre-COVID-19 period [ 25 ]. In addition to these studies, a review including 30 studies demonstrated differences in trends of different MDR bacteria during the pandemic [ 26 ]: the proportions of ESBL-producing E. coli and K. pneumoniae and carbapenem-resistant Pseudomonas aeruginosa (CRPA) decreased in most studies, whereas the proportions of other MDR bacteria including carbapenem-resistant Enterobacteriaceae (CRE), carbapenem-resistant Acinetobacter baumannii (CRAB), methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant Enterococci (VRE) increased.
The decreasing trends in the proportions of ESBL-producing E. coli in blood E. coli isolates observed in this study are in line with the latest EARS-Net report for years 2018–2022, although EARS-Net reports 3GC-resistance proportion instead of ESBL proportion [ 18 , 22 ]. The population-weighted mean proportion of 3GC resistance among invasive E. coli isolates decreased by 22.8%: from 7.9% in 2019 to 6.1% in 2022 in Finland [ 22 ]. This was nearly triple compared to the mean decrease in European Union (EU) and European Economic Area (EEA) countries (8.3%, from 15.6 to 14.3%), as well as greater than in some other European countries with traditionally low rates of AMR: Norway, 6.5%, from 6.2% in 2019 to 5.8% in 2022; Sweden, 3.8%, from 7.8 to 7.5%; and Denmark, 12,0%, from 7.5 to 6.6%. Moreover, in contrast to what we observed in Finland, the lowest 3CG resistance proportions were encountered already in 2021 in these countries, after which the trend may have reversed. Interestingly, in the Netherlands, the proportion of 3GC resistance remained stable during the COVID-19 pandemic (7.5% in 2019 and 7.7% in 2022). A similar phenomenon has also been reported in the UK, which was not included in the latest EARS-Net reports, where 3GC E. coli resistance in BSIs remained relatively stable at 14.5% between 2018 and 2022 [ 27 ]. Notably, in one Nordic country, Iceland, the proportion of 3GC resistance actually increased by 40%, from 7.0% in 2019 to 9.8% in 2022 [ 22 ].
For urine E. coli isolates, during 2019–2022, decreasing trends for cefadroxil-resistant isolates (representing ESBL-producing isolates) and 3GC-resistant isolates have been reported in the national surveillance reports of Sweden and Denmark, respectively [ 15 , 17 ]. However, the relative changes in these proportions were again smaller than observed in our study in Finland. Moreover, the lowest proportions of these isolates were reported already in 2021, after which the trend may have reversed, contrasting the data from Finland (Sweden: from 6.2% in 2019 to 5.9% in 2021 and 6.2% in 2022; Denmark: at hospital level from 6.9 to 5.8% in 2021 and 6.2% in 2022 and at primary health care from 5.2 to 4.4% in 2021 and 4.8% in 2022). Of note, in Norway, the proportion of ESBL-producing isolates was not reported to decrease among urine E. coli isolates during 2019–2022 [ 16 ]. However, the proportion there remained very low (3.0% in 2019 and 3.8% in 2022).
In contrast to most previous studies and reports, we showed that, although the decreases in the proportions of ESBL-producing E. coli were similar in most demographic groups and between sample types, the decreases in the incidence differed, reflecting the changes in tested isolates during the study period. The observed decrease in the total annual number of urine isolates tested and the resulting decrease in the number of E. coli isolates might reflect changes in diagnostic activity of UTIs or healthcare service access after the onset of the pandemic. Hence, particularly uncomplicated and/or non-severe UTIs may have been underdiagnosed during the pandemic years. Due to this selection bias, the proportion of ESBL-producing E. coli among urine E. coli isolates may be slightly overestimated, and the actual annual decrease may have been even larger. In addition, the reduction in elective care in hospitals may have decreased routinely sampled urine cultures, further affecting the numbers and proportions. For blood isolates, the previously observed continuous increase in the annual numbers [ 19 ] nearly stopped. This raises a question whether BSIs were also underdiagnosed during the pandemic. Importantly, in both sample types, the annual testing patterns did not clearly change, and the proportion of E. coli as a causative agent of UTIs and BSIs remained similar to pre-pandemic period, 69.3% and 44.0% during 2008–2019 [ 19 ] and 71.0% and 46.1% during 2020–2022, respectively.
In the context of COVID-19 pandemic, several factors may have influenced the decreasing trends observed in this study [ 28 , 29 ]. First, restrictions in travel, in particular international travel, may have significantly decreased the acquisition and cross-border import of ESBL-producing E. coli in Finland [ 30 , 31 ]. The number of travellers in Finnish airports decreased dramatically from 1.9 million in February 2020 to less than half in March 2020 and to only 1% in April 2020 [ 32 ]. Thereafter, the annual number of travellers increased but was over 10 million less in 2022 (15.6 million) compared to the pre-pandemic year 2019 (26.3 million) [ 33 ]. Similar trends were seen in at Swedish, Danish, Norwegian, and Dutch airports [ 34 , 35 , 36 , 37 ]. The decreased import of ESBL-producing E. coli via travel likely leads also in reduced onward transmission within household members, which is known to occur in up to 12% of the cases [ 38 ]. Second, the selective pressure of antibacterials reduced during the study period. The total consumption of antibacterials for systemic use in Finland decreased by 14.9% from 2019 to 2022, which was the greatest decrease among EU countries during the pandemic (EU mean: -2,5%) [ 39 ]. In 2022, Finland was among the EU/EAA countries with the lowest antibacterial consumption. The decrease in antibacterial consumption has been related to more stringent hygiene measures in prevention of COVID-19, which also decreased the spread of other respiratory pathogens [ 40 , 41 ] and resulted in the decreased usage of antibacterials. Third, the IPC measures in the hospitals and LTCFs in response to the pandemic may have decreased the spread of ESBL-producing E. coli in the health care setting [ 42 ].
Our study is not without limitations. First, the results of one major Finnish laboratory were not reported to the Finres database for year 2022. However, similar trends in the proportions of ESBL-producing isolates among blood E. coli isolates were observed according to local statistics (personal communication, KRJ, 13th of February 2024). Second, we do not know to what extent different factors (international travel, antimicrobial use, and IPC measures) contributed to the decrease in the proportions of ESBL-producing isolates. Third, the Finres database did not include information about community- or healthcare origin of the isolates. However, decreases in the proportions of ESBL-producing isolates in E. coli UTIs and BSIs in all age groups and both sexes suggest that the decrease likely happened in all settings, the community, acute care hospitals, and LTCFs. Last, we do not know whether the clinical outcome of these infections has changed during the study period.
After the onset of the COVID-19 pandemic, the proportion of ESBL-producing E. coli in UTIs and BSIs caused by E. coli significantly decreased during 2019–2022. Simultaneously, the risk of these infections decreased in most age groups. Although decreasing trends were similar between most of the age groups, the decrease in risk was most conspicuous among people aged ≥ 60 years, particularly among those ≥ 80 years of age. Overall, our results suggests that the decrease likely happened concurrently in both the community and healthcare settings. We assume that the rapid and prominent decrease in international travel was a major contributing factor, accompanied by decreased antibiotic use and pandemic-related IPC measures. Therefore, informing travellers on the risk of MDR bacteria related to international travel, hygiene measures, and appropriate antimicrobial use is crucial and evaluation of infection control measures in healthcare settings could be beneficial, especially in long-term care. In quarters 3 and 4 of 2022, the decreasing trend in ESBL-producing E. coli appeared to stabilise in urine cultures and even started to increase in blood cultures. The future trends in these proportions in different sample types and demographic age groups may further inform about the causes of source attribution of ESBL-producing E. coli . Continuous monitoring of the situation is therefore necessary, and the factors contributing to this decrease require further investigation.
Data availability
The datasets analysed during the current study are mostly included in this published article and its supplementary material, and they are mostly available in the Finres database, https://sampo.thl.fi/pivot/prod/fi/finres/lite/fact_lite . Part of the data were used under license for the current study and so are not publicly available. These data are available from the Finnish Institute for Health and Welfare (THL) for reasonable request.
Abbreviations
Average annual decrease
Average annual increase
Antimicrobial resistance
Compatibility interval
Carbapenem-resistant Acinetobacter baumannii
Carbapenem-resistant Enterobacteriaceae
Carbapenem-resistant Pseudomonas aeruginosa
European Economic Area
Net - European Antimicrobial Resistance Surveillance Network
European Union
European Committee on Antimicrobial Susceptibility Testing
Extended-spectrum β-lactamase
Infection prevention and control
Long-term care facility
Multidrug-resistant
Methicillin-resistant Staphylococcus aureus
Vancomycin-resistant Enterococci
Third-generation cephalosporins
Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–55. https://doi.org/10.1016/S0140-6736(21)02724-0 .
Article CAS Google Scholar
O´Neill J. Tackling drug-resistant infections globally: Final report and recommendations. The Wellcome Trust and the UK Department of Health. 2016. https://amr-review.org .
World Health Organization (WHO). Antibiotic resistance. https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance . Accessed 11 Dec 2023.
Salam MA, Al-Amin MY, Salam MT, Pawar JS, Akhter N, Rabaan AA, et al. Antimicrobial Resistance: a growing serious threat for Global Public Health. Healthc. 2023;11:1–20. https://doi.org/10.3390/healthcare11131946 .
Article Google Scholar
Rodríguez-Baño J, Rossolini GM, Schultsz C, Tacconelli E, Murthy S, Ohmagari N, et al. Antimicrobial resistance research in a post-pandemic world: insights on antimicrobial resistance research in the COVID-19 pandemic. J Glob Antimicrob Resist. 2021;25:5–7. https://doi.org/10.1016/j.jgar.2021.02.013 .
Article CAS PubMed PubMed Central Google Scholar
Getahun H, Smith I, Trivedi K, Paulin S, Balkhy HH. Tackling antimicrobial resistance in the COVID-19 pandemic. Bull World Health Organ. 2020;98:442–A442. https://doi.org/10.2471/BLT.20.268573 .
Article PubMed PubMed Central Google Scholar
Knight GM, Glover RE, McQuaid CF, Olaru ID, Gallandat K, Leclerc QJ, et al. Antimicrobial resistance and covid-19: intersections and implications. Elife. 2021;10:1–27. https://doi.org/10.7554/eLife.64139 .
Bonten M, Johnson JR, Van Den Biggelaar AHJ, Georgalis L, Geurtsen J, De Palacios PI, et al. Epidemiology of Escherichia coli Bacteremia: a systematic literature review. Clin Infect Dis. 2021;72:1211–9. https://doi.org/10.1093/cid/ciaa210 .
Article PubMed Google Scholar
MacKinnon MC, McEwen SA, Pearl DL, Lyytikäinen O, Jacobsson G, Collignon P, et al. Increasing incidence and antimicrobial resistance in Escherichia coli bloodstream infections: a multinational population-based cohort study. Antimicrob Resist Infect Control. 2021;10:131. https://doi.org/10.1186/s13756-021-00999-4 .
Kontula KSK, Skogberg K, Ollgren J, Järvinen A, Lyytikäinen O. Population-based study of Bloodstream Infection Incidence and mortality rates, Finland, 2004–2018. Emerg Infect Dis. 2021;27:2560. https://doi.org/10.3201/EID2710.204826 .
Laupland KB, Pasquill K, Steele L, Parfitt EC. Burden of bloodstream infection in older persons: a population-based study. BMC Geriatr. 2021;21:1–7. https://doi.org/10.1186/s12877-020-01984-z .
European Centre for Disease Prevention and Control (ECDC). Assessing the health burden of infections with antibiotic-resistant bacteria in the EU/EEA, 2016–2020. Stockholm: ECDC. 2022. https://www.ecdc.europa.eu/en/publications-data/health-burden-infections-antibiotic-resistant-bacteria-2016-2020 . Accessed 12 Dec 2023.
MacKinnon MC, McEwen SA, Pearl DL, Lyytikäinen O, Jacobsson G, Collignon P, et al. Mortality in Escherichia coli bloodstream infections: a multinational population-based cohort study. BMC Infect Dis. 2021;21:606. https://doi.org/10.1186/s12879-021-06326-x .
Ahava M, Ilmavirta H, Hakanen A, Salmenlinna S, Gunell M, Aittoniemi J et al. Bakteerien mikrobilääkeresistenssi Suomessa - Finres 2022. [Finnish Antimicrobial resistance report - Finres 2022]. Helsinki: Finnish Institute for Health and Welfare (THL); 2023. Finnish. https://urn.fi/URN:ISBN:978-952-408-187-0 . Accessed 11 Dec 2023.
National Veterinary Institute (SVA). Swedres-Svarm 2022. Sales of antibiotics and occurrence of resistance in Sweden. Solna/Uppsala; 2022. https://www.sva.se/en/our-topics/antibiotics/svarm-resistance-monitoring/swedres-svarm-reports . Accessed 11 Dec 2023.
NORM/NORM-VET. 2022. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Tromsø / Oslo; 2023. https://www.antibiotikaresistens.no . Accessed 11 Dec 2023.
Statens Serum Institut (SSI). DANMAP 2022 - Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Copengahen: SSI. 2022. https://www.danmap.org/ . Accessed 11 Dec 2023.
European Centre for Disease Prevention and Control (ECDC). Antimicrobial resistance in the EU/EEA (EARS-Net) - Annual Epidemiological Report 2022. Stockholm: ECDC. 2023. https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2022 . Accessed 11 Dec 2023.
Ilmavirta H, Ollgren J, Räisänen K, Kinnunen T, Hakanen AJ, Jalava J, et al. Increasing proportions of extended-spectrum β-lactamase-producing isolates among Escherichia coli from urine and bloodstream infections: results from a nationwide surveillance network, Finland, 2008 to 2019. Eurosurveillance. 2023;28. https://doi.org/10.2807/1560-7917.ES.2023.28.43.2200934 .
Finnish Institute for Health and Welfare (THL). Finres kuutio (julkinen). [Finres cube (public)]. Finnish. https://sampo.thl.fi/pivot/prod/fi/finres/lite/fact_lite . Accessed 29 Oct 2023.
European Committee on Antimicrobial Susceptibility Testing (EUCAST). Växjö: EUCAST. https://www.eucast.org/ . Accessed 23 Oct 2023.
European Centre for Disease Prevention and Control (ECDC). Country summaries – antimicrobial resistance in EU/EAA 2022. Stockholm: ECDC. 2023. https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2022 . Accessed 12 Dec 2023.
Lemenand O, Coeffic T, Thibaut S, Colomb Cotinat M, Caillon J, Birgand G. Decreasing proportion of extended-spectrum beta-lactamase among E. Coli infections during the COVID-19 pandemic in France. J Infect. 2021;83:664–70. https://doi.org/10.1016/j.jinf.2021.09.016 .
Article CAS PubMed Google Scholar
Hasan MR, Vincent YM, Leto D, Almohri H. Trends in the rates of extended-Spectrum-β-Lactamase-producing enterobacterales isolated from urine cultures during the COVID-19 pandemic in Ontario, Canada. Microbiol Spectr. 2023;11:1–12. https://doi.org/10.1128/spectrum.03124-22 .
Kuil K, Der W, Wielders CCH, Zwittink RD, de Greeff SC, Dongelmans DA, Kuijper EJ. Impact of the COVID-19 pandemic on prevalence of highly resistant microorganisms in hospitalised patients in the Netherlands, March 2020 to August 2022. Eurosurveillance. 2023;28. https://doi.org/10.2807/1560-7917.ES.2023.28.50.2300152 .
Abubakar U, Al-Anazi M, alanazi Z, Rodríguez-Baño J. Impact of COVID-19 pandemic on multidrug resistant gram positive and gram negative pathogens: a systematic review. J Infect Public Health. 2023;16:320–31. https://doi.org/10.1016/j.jiph.2022.12.022 .
UK Health Security Agency. English surveillance programme for antimicrobial utilisation and resistance (ESPAUR) Report 2022 to 2023. London: UK Health Security Agency. 2023. https://www.gov.uk/government/publications/english-surveillance-programme-antimicrobial-utilisation-and-resistance-espaur-report . Accessed 12 Dec 2023.
Monnet DL, Harbarth S. Will coronavirus disease (COVID-19) have an impact on antimicrobial resistance? Eurosurveillance. 2020;25:1–6. https://doi.org/10.2807/1560-7917.ES.2020.25.45.2001886 .
Seneghini M, Rüfenacht S, Babouee-Flury B, Flury D, Schlegel M, Kuster SP, et al. It is complicated: potential short- and long-term impact of coronavirus disease 2019 (COVID-19) on antimicrobial resistance - an expert review. Antimicrob Steward Healthc Epidemiol. 2022;2:1–8. https://doi.org/10.1017/ash.2022.10 .
Kantele A, Lääveri T, Mero S, Vilkman K, Pakkanen SH, Ollgren J, et al. Antimicrobials increase travelers’ risk of colonization by extended-spectrum betalactamase-producing enterobacteriaceae. Clin Infect Dis. 2015;60:837–46. https://doi.org/10.1093/cid/ciu957 .
Frost I, Van Boeckel TP, Pires J, Craig J, Laxminarayan R. Global geographic trends in antimicrobial resistance: the role of international travel. J Travel Med. 2019;26:1–13. https://doi.org/10.1093/jtm/taz036 .
Official Statistics of Finland (OSF): Air transport [e-publication]. Helsinki: Statistics Finland. 2020. https://www.stat.fi/til/ilma/2020/04/ilma_2020_04_2020-05-28_tie_001_en.html . Accessed 19 Dec 2023.
Statistics Finland. Air transport, 12ib -- Number of passengers and cargo tonnes at domestic airports by month, 2019M01-2024M01. Helsinki: Statistics Finland. https://pxdata.stat.fi/PxWeb/pxweb/en/StatFin/StatFin__ilma/statfin_ilma_pxt_12ib.px/ . Accessed 1 Feb 2024.
Swedish transport agency (Transportstyrelsen). Flygplatsstatistik. [Airport statistics]. Swedish. https://www.transportstyrelsen.se/sv/luftfart/statistik/Flygplatsstatistik-/ . Accessed 1 Feb 2024.
Statistics Netherlands. Aviation; monthly figures of Dutch airports. https://www.cbs.nl/en-gb/figures/detail/37478ENG . Accessed 6 Feb 2024.
Statistics Denmark [Internet]. Passengers on bigger public, manned Danish airports by airport and category of passenger - StatBank Denmark - data and statistics. https://www.statbank.dk/statbank5a/SelectVarVal/Define.asp?MainTable=FLYV31&PLanguage=1&PXSId=0&wsid=cftree . Accessed 1 Feb 2024.
Statistics Norway. 08507: Air transport - Passengers, by airport, traffic type and domestic/international flights 2009M01 - 2024M05. https://www.ssb.no/en/statbank/table/08507/ . Accessed 1 Feb 2024.
Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MCJ, van Genderen PJJ, Goorhuis A, et al. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis. 2017;17:78–85. https://doi.org/10.1016/S1473-3099(16)30319-X .
European Centre for Disease Prevention and Control (ECDC). Antimicrobial consumption in the EU / EEA (ESAC-Net) - Annual Epidemiological Report 2022. Stockholm: ECDC. 2023. https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-consumption-europe-2022 . Accessed 9 Jan 2024.
Hirae K, Hoshina T, Koga H. Impact of the COVID-19 pandemic on the epidemiology of other communicable diseases in Japan. Int J Infect Dis. 2023;128:265–71. https://doi.org/10.1016/j.ijid.2023.01.013 .
Shaw D, Abad R, Amin-Chowdhury Z, Bautista A, Bennett D, Broughton K, et al. Trends in invasive bacterial diseases during the first 2 years of the COVID-19 pandemic: analyses of prospective surveillance data from 30 countries and territories in the IRIS Consortium. Lancet Digit Heal. 2023;5:e582–93. https://doi.org/10.1016/S2589-7500(23)00108-5 .
European Centre for Disease Prevention and Control (ECDC). Systematic review of the effectiveness of infection control measures to prevent the transmission of extended-spectrum beta-lactamase-producing Enterobacteriaceae through cross-border transfer of patients. Stockholm: ECDC. 2014. https://www.ecdc.europa.eu/en/publications-data/systematic-review-effectiveness-infection-control-measures-prevent-transmission-0 . Accessed 9 Jan 2024.
Download references

Acknowledgements
Collaborative groups: We would like to thank all Finnish clinical microbiology laboratories and the corresponding clinical microbiology specialists for supplying susceptibility data to the national Finres database: Finnish Clinical Microbiology, FiRe laboratories (2023): HUS Diagnostic Center/Kymenlaakso Central Hospital, clinical microbiology (Merja Rautio), HUS Diagnostic Center/South Karelia Central Hospital, clinical microbiology (Merja Rautio), HUS Diagnostic Center/Bacteriology (Merja Rautio), Fimlab/Hämeenlinna; Fimlab/Jyväskylä (Dominik Kerimov), Fimlab/Lahti (Dominik Kerimov), Fimlab/Tampere (Dominik Kerimov), Fimlab/Vaasa (Emilia Lönnqvist), ISLAB Laboratory Centre/Joensuu (Jari Karhukorpi), ISLAB Laboratory Centre/Kuopio (Anne-Mari Rissanen and Heikki Ilmavirta), ISLAB Laboratory Centre/Southern Savonia (Päivi Suomala and Terhi Tuhkalainen), NordLab (Sini Koivunen and Joanna Peltola), SataDiag (Jari Kauranen), Seinäjoki Central Hospital (Jaana Kauppila), SYNLAB Finland, THL Mycobacterium Laboratory (Hanne-Leena Hyyryläinen, Silja Mentula), Tyks Laboratories, and Vita Laboratories (Vesa Kirjavainen, Päivi Mähönen). FiRe Steering Committee (2023): Kaisu Rantakokko-Jalava (chair, Tyks Laboratories), Kati Räisänen (THL), Nathalie Friberg (HUS Diagnostic Center), Hanne-Leena Hyyryläinen (THL), Heikki Ilmavirta (ISLAB Laboratory Centre), Jaana Kauppila (Seinäjoki Central Hospital), Sini Koivunen (Nordlab), Päivi Mähönen (Vita Laboratories), Anu Pätäri-Sampo (HUS Diagnostic Center), Tapio Seiskari (Fimlab), and Antti Hakanen (Tyks Laboratories).
No external funding was received for this study.
Author information
Authors and affiliations.
Department of Clinical Microbiology, Institute of Clinical Medicine, University of Eastern Finland, Kuopio, Finland
Heikki Ilmavirta & Tuure Kinnunen
ISLAB Laboratory Centre, Kuopio, Finland
Department of Health Security, Finnish Institute for Health and Welfare (THL), Helsinki, Finland
Heikki Ilmavirta, Jukka Ollgren, Kati Räisänen, Jari Jalava & Outi Lyytikäinen
Tyks Laboratories, Turku University Hospital (TYKS) and University of Turku (UTU), Turku, Finland
Antti Juhani Hakanen & Kaisu Rantakokko-Jalava
You can also search for this author in PubMed Google Scholar
Contributions
All the authors have significantly contributed to the study design and/or conduct. H.I., K.R., T.K., A.J.H., K.R-J., J.J. and O.L. drafted the manuscript and J.O. performed statistical analyses. All authors reviewed and contributed to the manuscript and read and approved the final manuscript.
Corresponding author
Correspondence to Heikki Ilmavirta .
Ethics declarations
Ethics approval and consent to participate.
According to the Research Act of Finland, the statement of the Regional Medical Research Ethics Committee was not required for the current study, since the study was register-based and it did not interfere with the integrity of the research subjects. An appropriate approval for the study, the study data, and the deriving publications was obtained from the Finnish Institute for Health and Welfare, the owner of the Finres database.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary material 2, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Ilmavirta, H., Ollgren, J., Räisänen, K. et al. Impact of the COVID-19 pandemic on extended-spectrum β-lactamase producing Escherichia coli in urinary tract and blood stream infections: results from a nationwide surveillance network, Finland, 2018 to 2022. Antimicrob Resist Infect Control 13 , 72 (2024). https://doi.org/10.1186/s13756-024-01427-z
Download citation
Received : 11 March 2024
Accepted : 22 June 2024
Published : 06 July 2024
DOI : https://doi.org/10.1186/s13756-024-01427-z
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Escherichia coli
- Urinary tract infection
- Bloodstream infection
- Decreased resistance
Antimicrobial Resistance & Infection Control
ISSN: 2047-2994
- General enquiries: [email protected]

Want to create or adapt books like this? Learn more about how Pressbooks supports open publishing practices.
Case Study #11: Jack in the Box E. Coli Crisis
On January 15, 1993, the Washington State Health Department alerted Robert Nugent, president of Jack in the Box, that the E. coli outbreak they had been informed of two days earlier, was at least partly attributed to hamburgers purchased at Jack in the Box restaurants (Sellnow and Ulmer, 1995). “Within a month, three children in the Seattle area, all under three, died of E. coli 0157:H7 poisoning – the strain linked to Jack in the Box” (p. 138).
One child had eaten at Jack in the Box, it was thought another was infected by a child who became ill after eating at Jack in the Box, and a cause for the third child’s infection was unknown. In total, 400 people were infected with the bacteria in Washington State, Idaho and Nevada. “As a result of this crisis, the Jack in the Box fast-food chain was not only in danger of losing sales, the company’s very existence was threatened by the crisis as well” (p. 138).
Primary Evidence
From the beginning of the crisis, Jack in the Box emphasized they were not solely responsible for the outbreak, pointing to the fact that customers not only ate at Jack in the Box, but other establishments as well. Although not accepting blame for the crisis, Jack in the Box tried to bolster their credibility by announcing in their first press release, January 18, 1993, they had taken measures to ensure all their menu items were prepared in accordance with an advisory issued by the Washington State Department of Health (Sellnow and Ulmer, 1995).
It wasn’t until January 21 that Jack in the Box took some responsibility for the crisis by announcing that the source of the problem was, in fact, contaminated meat. They explained they were reluctant to speculate before results from state tests came back which now indicated the problem was due to contaminated hamburger. Jack in the Box now pointed the finger at their meat supplier.
Robert Nugent also pointed the finger at the Washington Health Department and their apparent lack of passing out information in regards to new regulations, he also addressed corrective action the company was taking. His January 21, 1993 memo announced Jack in the Box would increase cooking times so the internal temperature of all hamburger products would exceed the new state regulations, check all grills to insure proper operating temperatures, and retrain all grill personnel on proper procedures.
Jack in the Box, on January 22, 1993, pledged “to do everything that is morally right for those individuals who had experienced illness after eating at Jack in the Box restaurants as well as their families” (Sellnow and Ulmer, 1995, p. 146). Jack in the Box dropped their criticism of the Washington State Health Department’s information distribution procedures February 12, 1993, and further emphasized their explanation of corrective measures (Sellnow and Ulmer, 1995).
Waiting a week to talk with the media is what really hurt the company’s reputation. “At the time I thought they were being unfair,” says Nugent. “It seemed to me they were more interested in placing blame than in really understanding what happened here” (p. 157). The story was prevalent for weeks, filled with accounts of organ damage and hospitalized kids. Foodmaker did not comment. “We had developed an attitude about PR that was something like, ‘Keep our mouth shut and if you want to talk with the press, have them call us,'” recalls Nugent (p. 157). Faced with negative publicity for a month, Robert Nugent replaced his public relations firm with President Jimmy Carter’s former press secretary, Joseph (Jody) Powell, who helped turn things around.
Secondary Evidence
Although officials at Foodmaker Inc., Jack in the Box’s parent company, claimed they first learned of the potential contamination on January 17, their initial response to destroy 20,000 pounds of potentially contaminated meat; to switch meat suppliers; to set up a toll-free number for complaints; and to raise cooking temperatures was seen as a positive move. (Soeder, 1993).
Not so positive though was the fact that they didn’t publicly accept responsibility for the food poisonings until the crisis was nearly a week old, and they partially blamed suppliers and state health officials. They also sought full recovery of losses and damages from their meat supplier, Vons. In response, the supplier issued this statement: “While we expected Foodmaker to sue its suppliers, we continue to be confident that Vons processing did not contaminate the meat. Health authorities have made it clear that proper cooking would have prevented this tragedy.” (Soeder, 1993).
According to Goff, (1999) “Foodmaker did the right things and did them swiftly…But when it came to communicating with the public, Nugent proved amazingly inept” (p. 157). Jack in the Box immediately suspended their hamburger sales, recalled meat from distributors, increased cooking times and temperatures, and pledged to pay all medical costs related to the disaster. Jack in the Box also hired a Dr. David Theno, a prominent food-safety consultant, to design an entirely new food-handling system (Goff, 1999).
Scholarly Journals
Jack in the Box, although still not taking responsibility for the crisis, was able to bolster their public image by emphasizing their willingness to alter the cooking procedures used in their restaurants and insisting their role in the crisis was speculative. (Sellnow and Ulmer, 1995). They emphasized the fact that they were taking actions to improve safeguards, while insisting that the crises was system-wide rather than specific to their organization. Although they insisted their products were not the source of many infections for which they were suspected, they offered to pay hospital bills of those who had eaten at their restaurants (Sellnow and Ulmer, 1995).
January 21, 1993, marked the day when Jack in the Box took some responsibility for the crisis. In a prepared statement by Robert Nugent, he addressed the contaminated meat and cooking temperatures, but also managed to shift the blame away from Jack in the Box. He explained that their investigation “traced the contaminated hamburger to a single supplier (Sellnow & Ulmer, 1995). “He also explained that the company had taken the ‘extraordinary step’ of replacing all hamburger patties in every restaurant in Washington and Idaho – despite the fact that health officials indicated this step was unnecessary” (p. 143).
Despite the fact Jack in the Box generated the argument of denial, the fact remained that their product had resulted in multiple deaths, and the public was still very skeptical. When Jack in the Box focused on external problems that contributed to the crisis, they tried to de-emphasize internal problems by focusing on the company’s history and their compliance with cooking regulations. “In fact, the history of our company’s compliance with those regulations is verified through numerous evaluations conducted by federal, state and local governments” (Ulmer & Sellnow, 2000, p. 151). In response to Jack in the Box’s failure to meet higher temperature guidelines imposed by the state, Nugent said the message had not reached his office. If Jack in the Box grills had been at the higher state standard, it is unlikely that the crisis would have occurred (Ulmer & Sellnow, 2000).
The Jack in the Box crisis falls under Coomb’s ‘accident’ category – “unintentional and happen during the course of normal organizational operations” (Coombs, 1995, p. 454). The crisis was devastating to Jack in the Box in the short-term. They had projected losses of between $20 and $30 million by March 24, 1993, resulting from the E. coli crisis. However, by the end of the year, they were able to slow their losses substantially.
Jack in the Box used several strategies to weather the E. coli storm. They began with a combination of avoidance and attachment strategies. When they issued their first press release explaining the source of the illness was unclear; and that some, but not all of the people being treated had eaten at their restaurant they employed two types of avoidance strategies – denial of intention and denial of volition. They also used an attachment strategy – bolstering – in the same press release when they announced the extra measures they were taking to ensure all food was cooked in accordance with a new state advisory that was issued.
They also employed scapegoating, a form of denial of volition (avoidance strategy) when they first took some responsibility for the crisis January 21, but blamed their meat supplier for the contaminated meat. They also used forgiveness strategies, more specifically remediation, when they announced they would pay all the medical bills for people who became ill during the E. coli crisis. Another forgiveness strategy, rectification, was used when Jack in the Box announced several corrective actions they were taking in regards to cooking temperatures.
Although Foodmaker, Jack in the Box’s parent company, continued to be in the news for years following the crisis – every time E. coli came up the whole Jack in the Box story would resurface – the company survived. By referring reporters to articles regarding Foodmaker’s food-safety innovations, the company regained credibility. In 1994, they instituted the fast-food industry’s first comprehensive food-safety program, the Hazard Analysis & Critical Control Points system. Today, they are considered the leader in food safety in the fast-food industry (Liddle, 1997) and they are the country’s fifth-largest burger chain (Goff, 1999).
Attributions
The document to which this notice is attached is protected by copyright owned in whole or in principal part by the University of Oklahoma Board of Regents and the state of Oklahoma. You may download the document for reference and research purposes only.
Distribution and/or alteration by not-for-profit research or educational institutions for their local use is permitted as long as this notice is kept intact and attached to the document. Any other distribution of copies of the document or any altered version there of is expressly prohibited without prior written consent of the University.
For permission requests, licensing proposals, questions, or further information, please contact Candace Stimmel .
Case Study #11: Jack in the Box E. Coli Crisis Copyright © by Sam Schechter is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License , except where otherwise noted.
Share This Book
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager
Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection
Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
A fast and simple approach to optimize the unit operation high pressure homogenization - a case study for a soluble therapeutic protein in E. coli
Affiliations.
- 1 a Research Division Biochemical Engineering , Institute of Chemical, Environmental and Bioscience Engineering, TU Wien , Vienna , Austria.
- 2 b Christian Doppler Laboratory for Mechanistic and Physiological Methods for Improved Bioprocesses , Institute of Chemical, Environmental and Bioscience Engineering, TU Wien , Vienna , Austria.
- PMID: 30664394
- DOI: 10.1080/10826068.2018.1536988
Escherichia coli is one of the most commonly used host organisms for the production of recombinant biopharmaceuticals. E. coli is usually characterized by fast growth on cheap media and high productivity, but one drawback is its intracellular product formation. Product recovery from E. coli bioprocesses requires tedious downstream processing (DSP). A typical E. coli DSP for an intracellular product starts with a cell disruption step to access the product. Different methods exist, but a scalable process is usually achieved by high pressure homogenization (HPH). The protocols for HPH are often applied universally without adapting them to the recombinant product, even though HPH can affect product quantity and quality. Based on our previous study on cell disruption efficiency, we aimed at screening operational conditions to maximize not only product quantity, but also product quality of a soluble therapeutic protein expressed in E. coli. We screened for critical process parameters (CPPs) using a multivariate approach (design of experiments; DoE) during HPH to maximize product titer and achieve sufficient product quality, based on predefined critical quality attributes (CQAs). In this case study, we were able to gain valuable knowledge on the efficiency of HPH on E. coli cell disruption, product release and its impact on CQAs. Our results show that HPH is a key unit operation that has to be optimized for each product.
Keywords: Critical quality attribute; downstream processing;; high pressure homogenization; process optimization; soluble protein.
PubMed Disclaimer
Similar articles
- High Pressure Homogenization for Inclusion Body Isolation. Ebner J, Sedlmayr V, Klausser R. Ebner J, et al. Methods Mol Biol. 2023;2617:141-154. doi: 10.1007/978-1-0716-2930-7_9. Methods Mol Biol. 2023. PMID: 36656521
- Genetic engineering modification and fermentation optimization for extracellular production of recombinant proteins using Escherichia coli. Zhou Y, Lu Z, Wang X, Selvaraj JN, Zhang G. Zhou Y, et al. Appl Microbiol Biotechnol. 2018 Feb;102(4):1545-1556. doi: 10.1007/s00253-017-8700-z. Epub 2017 Dec 21. Appl Microbiol Biotechnol. 2018. PMID: 29270732 Review.
- A combination of HPLC and automated data analysis for monitoring the efficiency of high-pressure homogenization. Eggenreich B, Rajamanickam V, Wurm DJ, Fricke J, Herwig C, Spadiut O. Eggenreich B, et al. Microb Cell Fact. 2017 Aug 1;16(1):134. doi: 10.1186/s12934-017-0749-y. Microb Cell Fact. 2017. PMID: 28764719 Free PMC article.
- A novel method to recover inclusion body protein from recombinant E. coli fed-batch processes based on phage ΦX174-derived lysis protein E. Ehgartner D, Sagmeister P, Langemann T, Meitz A, Lubitz W, Herwig C. Ehgartner D, et al. Appl Microbiol Biotechnol. 2017 Jul;101(14):5603-5614. doi: 10.1007/s00253-017-8281-x. Epub 2017 Apr 20. Appl Microbiol Biotechnol. 2017. PMID: 28429059 Free PMC article.
- Purification of secreted recombinant proteins from Escherichia coli. Le HV, Trotta PP. Le HV, et al. Bioprocess Technol. 1991;12:163-81. Bioprocess Technol. 1991. PMID: 1367083 Review.
- Selective Release of Recombinant Periplasmic Protein From E. coli Using Continuous Pulsed Electric Field Treatment. Schottroff F, Kastenhofer J, Spadiut O, Jaeger H, Wurm DJ. Schottroff F, et al. Front Bioeng Biotechnol. 2021 Feb 9;8:586833. doi: 10.3389/fbioe.2020.586833. eCollection 2020. Front Bioeng Biotechnol. 2021. PMID: 33634078 Free PMC article.
- Recent Developments in Bioprocessing of Recombinant Proteins: Expression Hosts and Process Development. Tripathi NK, Shrivastava A. Tripathi NK, et al. Front Bioeng Biotechnol. 2019 Dec 20;7:420. doi: 10.3389/fbioe.2019.00420. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31921823 Free PMC article. Review.
- Search in MeSH
Related information
Linkout - more resources, full text sources.
- Taylor & Francis
Miscellaneous
- NCI CPTAC Assay Portal
- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Infect Drug Resist
E. coli O157 outbreaks in the United Kingdom: past, present, and future
Thomas hugh pennington.
University of Aberdeen, Aberdeen, United Kingdom
This review describes Escherichia coli O157 outbreaks in the United Kingdom, beginning from the first, in the 1980s, to those recorded in 2013. We point out that the United Kingdom differs from other countries, particularly the United States, in that it has had a considerable number of outbreaks associated with butchers, but very few caused by contaminated burgers. Two of the butcher-associated outbreaks (in central Scotland in 1996 and South Wales in 2005) were very large and are considered here in detail; the reviewer conducted detailed investigations into both outbreaks. Also considered is the very large outbreak that occurred in visitors to an open farm in Surrey in 2009. Detailed descriptions of some milk-borne outbreaks and incidents connected with camping and childrens’ nurseries have been published, and these are also considered in this review. Large outbreaks in the United Kingdom have sometimes led to policy developments regarding food safety, and these are considered, together with public reactions to them, their health effect, and their value, as examples to follow or eschew in terms of the procedures to be adopted in response to incidents of this kind. Regulatory and legal consequences are also considered. As a wise man said, making predictions is difficult, particularly about the future. This review follows this position but points out that although human infections caused by E. coli O157 are rare in the United Kingdom, their incidence has not changed significantly in the last 17 years. This review points out that although a response to an outbreak is to say “lessons must be learned”, this response has been tempered by forgetfulness. Accordingly, this review restricts its recommendations regarding outbreaks to two: the crucial importance of a rapid response and the importance of experience, and even “gut feeling”, when an inspector is evaluating the safety of a food business.
Introduction
Theodor Escherich in Munich in 1885 was the first to grow Escherichia coli in pure culture. He also showed that it was a normal inhabitant of the intestines of infants. 1 About 100 billion billion E. coli are alive in the world at any one time, and the overwhelming majority are harmless commensals. However, some are pathogenic. This review considers outbreaks of disease in the United Kingdom caused by E. coli O157, one of the most important pathogens with this specific identity and a member of a group of organisms known variously as enterohemorrhagic, verocytotoxin-producing, or Shiga toxin-producing E. coli . 2
Most E. coli O157 strains produce Shiga toxin type 2; its B subunits bind to the surface of intestinal cells, and its A subunits enter and turn off protein synthesis by disrupting the large ribosomal subunit. Bloody diarrhea is a hallmark of symptomatic cases; a minority of patients, usually at the extremes of age, develop hemolytic uremic syndrome (HUS), in which thrombocytopenia and renal failure usually occur. Neurological and cardiac complications are important determinants of mortality. 3
Human infections are zoonotic; E. coli O157 is a normal nonpathogenic intestinal commensal in ruminants. 4
Outbreaks in the United Kingdom (generally defined as two or more cases from separate households linked to a common source) are investigated by outbreak control teams (OCTs). Factors that lead to the declaration of an outbreak and the establishment of an OCT are one or more of the following: an immediate and/or continuing health hazard to the local population, one or more cases of serious disease, large numbers of cases, and the involvement of more than one local authority. OCT members include public health specialists, local authority environmental health officers, and health service staff. Their main functions are to manage the outbreak, to draw lessons, and to prepare a report. Many reports only circulate locally; a few are published in the scientific literature. Occasionally, accounts are published as the outcome of nonstatutory inquiries commissioned by an interested party, such as the review of the major outbreak of E. coli O157 in Surrey in 2009 (the Godstone Farm outbreak). 5 Statutory inquisitorial inquiries include fatal accident inquiries (Scotland only) and public inquiries. Prosecutions in the United Kingdom are adversarial.
The first E. coli O157 outbreaks
Outbreaks of bloody diarrhea in 1982 in the United States marked the sudden appearance of E. coli O157 as a new pathogen, with 25 cases in Medford, Oregon, in February and March, and 18 cases in Traverse City, Michigan, in May and June. The organism was isolated from four cases in each outbreak. There was a strong epidemiological link to the consumption of beef burgers. 6
E. coli O157 was first isolated during an outbreak in the United Kingdom in July 1983, when stools from three patients tested positive in a cluster of 11 cases of HUS in children from Wolverhampton in the West Midlands. 7 It was a pathogen new to the United Kingdom. A retrospective search of more than 15,000 E. coli strains collected between 1978 and 1982 revealed only one isolate, and it was not found in any of the 161 diarrhea outbreaks investigated by the Reference Laboratory of the Public Health Laboratory Service for England and Wales between 1973 and 1983. 6
Outbreaks in the United Kingdom, 1985–1996
The first community outbreak in adults in the United Kingdom occurred in July 1985, when 49 people fell ill in East Anglia, 19 of whom were admitted to hospital. Bloody diarrhea and severe abdominal pain dominated. Three patients had laparotomies, and one woman (aged 64 years) died from fulminant colitis. Thirty-eight cases were women. E. coli O157 was isolated from the stools of 24 cases; their mean age was 43 years. The investigators found no link with the consumption of beef burgers (three of the confirmed cases were vegetarians) but suspected that handling foods followed by hand-to-mouth transmission rather than food-borne infection had occurred. A case-control study showed a strong association between infection and the preparation of raw vegetables, especially potatoes. 8
In England and Wales, at least 55 outbreaks were reported between 1983 and 1996, and between 1989 and October 1996, there were 24 outbreaks in Scotland. 9 Microbiologically confirmed sources included animal contact at a farm visitor center and cheese. Particularly good data came from Wales, which in 1990 had introduced universal testing for E. coli O157 for all first-time acute-phase fecal specimens. 10 By 1998, 415 cases had been reported, 46.3% with blood in their stools. The majority of cases (82.2%) did not occur in outbreaks. Six outbreaks occurred during this period: four with the mode of spread being person to person (two in institutions caring for psychogeriatric patients and two in day nurseries) and one being spread by contaminated meat.
Heuristically important outbreaks in Scotland in 1990 and 1994
The Hartwoodhill Hospital outbreak occurred in October 1990. 11 Eight patients and three staff members in this small psychogeriatric hospital in Lanarkshire were infected. Four patients died. A fatal accident inquiry was held in 1991. This process requires a sheriff (a district judge) to make a determination in respect of a death, “setting out the circumstances of the death as far as they have been established to his satisfaction”, including “the reasonable precautions, if any, whereby the death and any accident resulting in the death might have been avoided, and the defects, if any, in any system of working which contributed to the death or any accident resulting in the death”. The sheriff concluded that an infection control nurse “could reasonably have been expected to ensure that the justifiable criticism of lax hygiene in the uplifting of soiled laundry, of unsafe standards of cleanliness in the wards and their furnishings, and of poor standards of housekeeping in the washing up of cleaning bowls and utensils would not have been levied”.
The West Lothian (Redhouse Dairy) milk-borne outbreak occurred in May 1994. 12 It affected a rural community to the west of Edinburgh. The dairy obtained milk from outlying feeder farms and pasteurized, bottled, and delivered it to more than 1,000 domestic customers and some retail outlets. More than 100 people were infected. One child died early in the outbreak, and 24 were admitted to hospital. Their average length of stay was just over 25 days. Ten children developed HUS, and six needed dialysis. Isolates of E. coli O157 were made from the stools of 69 patients, a section of pipe connecting the pasteurizer to the bottling apparatus, a bottling machine rubber, raw milk from a bulk carrier from a farm that supplied the dairy, and bovine feces from the same farm. All the isolates had the same pulsed field gel electrophoresis (PFGE) type, which was different from that of other strains being isolated in Scotland at that time. This influenced the sheriff at the criminal trial. Despite the defense taking the line that “there was not sufficient evidence of a credible and reliable nature to establish that milk sold or deposited for sale was contaminated with E. coli O157 organisms”, the accused were found guilty of selling contaminated milk. Not only was this one of the earliest outbreaks in the United Kingdom to be investigated by PFGE of isolates, it was the first recorded anywhere in the world to involve a heat-treated milk supply. Its heuristic importance also comes from it being the subject of a detailed economic and social assessment of its effect on the infected and their families, hospitals and primary and community care facilities, public health departments, laboratories, environmental health departments, and veterinary services not only during the outbreak but also for the following 12 months. 13 One year after the outbreak, one child had had a renal transplant and two still had no renal function and were receiving peritoneal dialysis; one of these children had developed insulin-dependent diabetes, and the other required intermittent nasogastric feeding. Two other children had reduced renal function. The medical, productivity loss, and outbreak control costs were estimated to be £3.2 million for the first year. Over the course of 30 years, the costs were projected to be £11.9 million. This outbreak ranks fourth in case numbers in the United Kingdom to date.
Other milk-borne outbreaks
The North Cumbria milk-borne outbreak ranks third in case numbers in the United Kingdom. 14 It affected 114 individuals in late February and early March 1999; 88 had E. coli O157 confirmed by laboratory tests. Twenty-eight were admitted to hospital, and three nursery school-age children developed HUS, recovering after hemodialysis. The milk came from a farm with 65 cows in milk. About 600 pints were pasteurized on Sundays, Tuesdays, and Thursdays and went to 321 premises, including 11 commercial establishments. The pasteurizer on the farm was of an old type that had been the subject of a food hazard warning the previous year. New heat exchanger plates had been fitted by the farmer a few days before the outbreak, but there was no record of subsequent tests. There was also failure in the automatic recording of flow diversion activity and inadequate temperature monitoring. It was likely that heat-treated milk was being contaminated with raw milk at the heat exchanger unit. Although milk samples did not grow E. coli O157, the organism was recovered from calf pen straw bedding, slurry samples, the floors of pens housing animals, and 11 animal feces samples. All these isolates had the same or very similar PFGE patterns as the human outbreak isolates. The farmer pleaded guilty when prosecuted by the local authority for the sale of milk unfit for consumption and for breaches of the regulations concerning the operation of pasteurizers.
During 1992–2000, E. coli O157 was the most common cause of milk-borne general outbreaks of infectious intestinal disease in England and Wales. 15 It was the causative agent in nine outbreaks; Campylobacter caused seven outbreaks, Salmonella typhimurium six, Salmonella enteritidis pt4 two, other salmonellas two, and Cryptosporidium one. Five of the E. coli O157 outbreaks were ascribed to the consumption of unpasteurized milk, one to pasteurized milk that had been mixed with unpasteurized milk, and three to milk sold as pasteurized. Small farm dairies that bottled their own milk were identified as a significant problem. Unlike large dairies, which conducted the alkaline phosphatase test on all batches, during this period, small on-farm dairies did not do daily tests.
The 1996 central Scotland outbreak
Outbreaks of intestinal infectious disease associated with butchers’ premises were not rare before 1996. Some were very big, such as the enormous North Wales and Cheshire outbreak during the summer of 1989. Caused by Salmonella typhimurium DT 12, it affected 640 people, with 74 hospital admissions and three deaths. 16 In 1995, there were eight butcher-associated outbreaks in England and Wales. 17
The central Scotland outbreak occurred in November and December 1996. The first evidence of infection was the identification of a presumptive E. coli O157 strain from a 5-year-old child with bloody diarrhea on November 21. 18 , 19 Two more provisional identifications were made the next day, when the identity of the first isolate was confirmed. The microbiologist had been involved in the Hartwoodhill outbreak. He immediately considered that another outbreak was underway, because he considered his laboratory to be in a low-risk area and he had been screening feces for E. coli O157 for several years and had only made three isolates in the previous four years. He made further investigations and found that two other patients had been admitted to his hospital with bloody diarrhea and that another two with the same problem were in another local hospital. All lived in Wishaw, a small town in Lanarkshire, near Glasgow. By evening, the local public health department had identified 15 confirmed or suspected cases with E. coli O157. Eight had eaten food from J Barr and Son, Butchers, of Wishaw or had attended a church lunch in Wishaw at which food had been provided by Barrs. The number of cases increased dramatically. Eventually there were 503 cases, 279 of which were confirmed microbiologically ( Figure 1 ). All isolates tested by PFGE had indistinguishable profiles. Cases were reported across central Scotland. The outbreak was made up of several separate incidents: the church lunch, a birthday party held in a public house on November 23, cases in a nursing home, and retail sales. Seventeen patients died directly of infection with the outbreak strain, and there were four associated deaths. The 74 attendees at the church lunch were the hardest hit ( Figure 2 ). The lunch was for the elderly: eight died, and six developed renal failure. Their ages ranged from 70 to 83 years. The outcomes of infection contracted at the pub party were very different. Most of the 129 attendees were young (only 12 were aged 40 years or older). Of the 25 patients who were infected, 11 tested positive but had no symptoms. E. coli O157 was isolated from 10 members of Barr’s staff, but only three had symptoms. The outbreak strain was isolated from the boiler used to cook the steak served at the church lunch; the butcher’s vacuum packing machine; raw sirloin, sausage stewing steak, and legs of pork from within the Barr premises; gravy from the church lunch; cooked ham supplied to the pub for the party; roast beef; roast pork; and corned beef supplied by Barr to other firms. The butcher pleaded guilty in January 1998 on charges under the Food Safety Act and was fined a total of £2,250.

Epidemic curve, Central Scotland 1996 outbreak.
Note: Data from Pennington. 20
Abbreviations: Nov, November; Dec, December.

Epidemic curve, Church Hall component of the Central Scotland outbreak.
The outbreak led to two inquiries
The government in Scotland, the Scottish Office, established an expert group, chaired by the author, on November 28, 1996. The outbreak was still in full progress. Its serious nature was already evident, in that five patients had already died. My group was charged “to examine the circumstances which lead to the outbreak … and to advise [the Secretary of State for Scotland] on the implications for food safety and the lessons to be learned”. Our final report was presented to parliament in April 1997. 20 It concluded that measures were needed to reinforce legislation to strengthen the implementation and enforcement of the Hazard Analysis and Critical Control Point System (HACCP). The HACCP is a structured approach to analyzing the potential hazards in an operation, identifying the points in the operation where the hazards may occur, and deciding which points are critical to control to ensure consumer safety. These critical control points are then monitored, and remedial action, specified in advance, is taken if conditions at any point are not within safe limits. Verification procedures are established to confirm that the HACCP system is working effectively, and documentation of all procedures and appropriate records is done. My group recommended that all these HACCP principles be adopted by all food businesses. While this was being negotiated into EU and domestic legislation, selective licensing arrangements for premises selling raw and unwrapped cooked meats should be introduced. Licensing would require food handler training and arrangements for ensuring the physical separation of the raw and the cooked. Within a year, local authorities in Great Britain received £19 million to be spent over the course of 3 years to facilitate the acceleration of the HACCP. Butcher’s licensing was introduced in Scotland and England in 2000, and in Wales and Northern Ireland in 2001, and remained in force until the beginning of 2006. Abattoirs were also considered. Recommendations to the Meat Hygiene Service included rigor in the rejection of dirty animals and the targeting of resources on higher-risk premises, particularly those with Hygiene Assessment scores below 65. 17
A fatal accident inquiry established the details of what went wrong at the Barr premises. 21 Set up in December 1996, the inquiry had to wait until criminal proceedings against John Barr had been concluded and did not begin until in April 1998. The sheriff made many criticisms of Barr’s. He listed deficiencies under the heading of “defects in any system of working which contributed to the accident”. Temperature probes were not being used to ensure the proper cooking of meat. Separate knives, work tables, scales, and vacuum packers for raw and cooked meats were not provided. There was no clear management structure to enforce food safety measures. And environmental health officers had failed to identify these food safety hazards. Detailed examples that the sheriff quoted were the defective boiler used to cook the meat served at the church lunch (two of its heating elements were not working), work flows in the premises allowing the crossing of raw and cooked meat processing, and the use of the same surfaces for handling raw and cooked meats (surfaces that were not being cleaned with bactericides).
What was being used was a biodegradable washing up liquid for cleaning work surfaces. The description ‘biodegradable’ in the eyes of Barr’s senior staff was synonymous with ‘bactericidal’. The liquid in use was green in color. There was no doubt about that. Mr Barr thought that about five years before the outbreak he had changed his supplier on the recommendation of a former employee who said that he could get a cleaning agent with the same properties at a more attractive price. If true this raises the question […] how environmental health officers in the course of their inspections did not discover that a bactericidal agent was not being used. [Sheriff]
Studies in my laboratory showed that the “green biodegradable liquid” supported the growth of E. coli. Not long before the outbreak, Mr Barr had been voted “Scottish butcher of the year” by his customers. The sheriff summarized the situation: “I have no doubt Mr John Barr liked a clean shop and maintained a clean shop. What he failed to do was maintain a safe shop and the main ingredients of his failure was ignorance of the requirements which would produce that result.” The outbreak occurred because of Barr’s ignorance; it was big because his business was big. Outwardly a small local butcher, the business had about 40 employees, and at the time of the outbreak, it had a substantial wholesale and retail trade involving the production and distribution across central Scotland of raw and cooked meats and bakery products.
The 2005 South Wales outbreak
General outbreaks of intestinal infectious disease associated with butchers’ premises continued in the United Kingdom after 1996. 17 In England and Wales, there were four outbreaks in 1997, two in 1998, four in 1999, five in 2000, three in 2001, and one in 2004. There were none in 2002 and 2003. However, the expectation that butchers licensing and its associated focus on the implementation of HACCP would result in the prevention of butcher-associated outbreaks was dashed in South Wales in 2005. 17 On September 16, a doctor at Prince Charles Hospital, Merthyr Tydfil, informed the National Public Health service for Wales that in the previous two days, five children had been admitted with bloody diarrhea and that three others with the same condition had been seen at the hospital assessment unit. That morning, microbiology reported that E. coli O157 had been isolated from two samples. An outbreak was declared that afternoon. At its end, 118 cases had been confirmed microbiologically and 39 were probable, in that the patients had developed bloody diarrhea during the outbreak period. The majority of cases were schoolchildren. Thirty-six primary and eight secondary schools in the local authority areas of Bridgend, Caerphilly, Merthyr Tydfil, and Rhondda Cynon Taf had at least one case; 25 schools had only one case, but two had the maximum recorded in the outbreak (eleven cases). Forty-eight cases were considered to be secondary infections ( Figure 3 ). Thirty-one cases were admitted to hospital, eight with HUS.

Epidemic curve, South Wales 2005 outbreak.
Notes: © Queen’s Printer and Controller of HMSO; 2009. Pennington TH. The Public Inquiry into the September 2005 Outbreak of E. coli O157 in South Wales . Aberdeen: HMSO; 2009. Available from: http://wales.gov.uk/ecolidocs/3008707/reporten.pdf?skip=1&lang=en . 17
Abbreviation: NPHS, Public Health Service for Wales.
One child died. Mason Jones was aged 5 years and had just started school. His illness began on September 21, with a fever. Diarrhea started on September 22, and a stool sample was taken. The diarrhea became bloody on September 23. Late on September 25, Mason was admitted to hospital. He had developed thrombocytopenia and had biochemical changes indicating the onset of renal failure. The stool sample result was positive for E. coli O157. Peritoneal dialysis started on September 26, but Mason had fits on September 27. He needed drugs to maintain his blood pressure and was being ventilated. Hemodialysis was started, and Mason’s condition stabilized. His best renal day was October 1, when hemodialysis was not required; however, it was required again on October 3. At 5 pm, his blood pressure began to fall, dropping suddenly at 11 pm. Some cardiac function returned after external cardiac massage, but at midnight, there was a precipitous fall in blood pressure and a gradual drop in his heart rate. He became unresponsive, and resuscitation attempts stopped at 12.30 am on October 4.
On October 5, the National Assembly for Wales set up a cross-party committee to consider the terms of reference for a public inquiry. It was formally established by the assembly under my chairmanship on December 7, 2005.
The common feature linking the cases was the supply of cooked, sliced meat from John Tudor and Son. The business supplied the school meals service in the four local authorities. All schools with a case where the onset of symptoms took place before September 17, 2005, had been exposed to cold cooked meats supplied by Tudors during the first week of term. Very similar PFGE profiles and indistinguishable variable number tandem repeat profiles were shown by all the strains tested from outbreak cases (including the one from Mason Jones), as well as strains isolated from unused cooked meat recovered from five schools, from a joint of raw meat recovered from John Tudor and Sons premises, and from cattle feces from the farm that supplied the abattoir that supplied meat to Tudors.
The abattoir was operated by Jonathan Tudor, a cousin of William Tudor, who ran the butcher’s business known as John Tudor and Son. The abattoir was built in about 1860. In 1994, a scheme for scoring and assessing hygiene at abattoirs was introduced. Marks were out of 100. Scores below 66 were deemed to be unacceptably low. In March, the Tudor abattoir scored 15 points. This score rose to 35 in July, but at an unannounced inspection in August, it had fallen to 11, the lowest score ever recorded in Great Britain. The government was advised to revoke the abattoir’s license, but it continued to operate. A HACCP for plants such as the abattoir was introduced in 2003, but at the time of the outbreak, it had not been implemented at the abattoir. In essence, the problems identified in the early 1990s had not been rectified; the regulator, the Meat Hygiene Service, had failed to perform its enforcement function effectively.
John Tudor and Son sold raw and cooked meats. It cooked meats and produced faggots and burgers. Public sector organizations were major customers. Investigation of the business after the outbreak found many serious food hygiene deficiencies: Staff were poorly trained; single machines and equipment, including weighing scales, were being used for both raw and cooked meats; and cleaning was totally inadequate (eg, the probe wipes found on the premises had an expiry date of 1991, and machinery was usually cleaned with “truck wash traffic film remover”). For some products, the HACCP plan described a cooling rate after cooking that flouted the laws of physics. The plan also did not cover a major activity at the premises: the processing of bought-in cooked meats. Most staff members had not seen it or knew what the HACCP meant. A forensic scientist produced conclusive evidence that temperature records were not made contemporaneously but were made in batches at one time. The longest period written by one person in one ink was from July 28, 2004, to February 2, 2005, which are significant dates because on July 28, 2004, the premises had been inspected regarding butchers licensing, and on February 2, 2005, there had been a follow-up inspection after a January 18, 2005, inspection because in January, William Tudor had been unable to produce the records, saying they had been “taken home for updating”. In saying this, he lied to environmental health officers. Even so, one environmental health officer noted in April 2001 that, “Records were being kept, systemic analysis, but couldn’t help wondering whether some records were fixed as same style writing and color pen on many of the records.” However, her note was not followed up by her successors. When meat went bad, William Tudor often directed his staff to reintroduce it into the food chain by removing the bad parts of a joint or by putting it into faggot mix, which hid the smell. In September 2007, he pleaded guilty to six offences of placing unsafe food on the market and one of failing to protect food against the risk of contamination. He was sentenced to 12 months imprisonment and prohibited from participating in the management of any food business in the future.
William Tudor’s premises were inspected regularly by local authority environmental health officers. Their job was difficult because of his dishonesty, but they also failed to pick up fundamental flaws in the HACCP plan and defects in its implementation. William Tudor was issued with a butcher’s license 6 weeks before the start of the outbreak. He should not have been.
My public inquiry report was published in March 2009. 17 It made 24 recommendations, 15 of which focused on the HACCP and related issues. In response, the Food Standards Agency set up its Food Hygiene Delivery Programme, a 4-year implementation scheme.
Outbreaks associated with farm visits/direct contact with manure
Twenty-three outbreaks of E. coli O157 were linked to visits to open/petting farms in England and Wales between 1994 and 2008. 22 The number of those affected in each outbreak averaged 7.7 cases. In Scotland, animal contact accounted for the largest single category of E. coli O157 outbreaks reported from 1996 to 2008.
A particularly tragic and costly outbreak occurred at an open farm in Hertfordshire, just north of London, in 1997. 22 The farm had about 112,000 visitors annually. Hand-washing facilities were provided at the touching barn, the classrooms, and the restaurant. A boy, aged 7 years, who lived on the farm and who mucked out calf pens and had free access to all parts developed bloody diarrhea on May 10. He spent one week in hospital, and E. coli O157 was isolated from his stools. In addition, a 6-year-old girl who had visited the farm on June 3 and touched a number of animals fell ill on the June 6 and was admitted to hospital on June 10 with diarrhea and vomiting. She developed HUS, recovering with conservative management. Her stools were positive for E. coli O157. On June 30, a 4-year-old boy developed bloody diarrhea. E. coli O157 was isolated from his stools that had the same PFGE pattern as the strains from the other cases and from three isolates made on the farm from a goat paddock and one from a cow. The boy had visited the farm on June 27 and had clambered on fences and stroked animals. He was admitted to hospital on July 2 and developed HUS with severe neurological complications. He was in a coma for 12 days and was left unable to speak or eat and with epilepsy and spastic quadriplegia. Legal proceedings started. In January 2001, a settlement of £2.6 million was agreed on. He died in April 2006.
A much larger outbreak occurred in Scotland in 2000. 23 A scout camp was held at the New Deer Agricultural Show Ground in Aberdeenshire to celebrate the Millennium. It was intended that the camp should run from Friday, May 26, to Sunday, May 28, but the camp was abandoned on May 27 because of very heavy rain. On June 1, a cub scout aged 8 years was admitted to the Royal Aberdeen Children’s Hospital with gastroenteritis. His stools tested positive for E. coli O157, and the result was sent to the local communicable disease team on June 2. His attendance at the camp was noted, and the Scout Association was asked to compile a list of attendees. On June 4, a second child was admitted to Royal Aberdeen Children’s Hospital with a probable diagnosis of E. coli O157. A public health doctor determined that other camp attendees were unwell, including an adolescent who had been admitted to the Infection Unit of Aberdeen Royal Infirmary with symptoms suggestive of an E. coli O157 infection. An outbreak was declared. The OCT was convened at 6 pm, and a press release was issued describing the scout camp as a potential link with cases of gastroenteritis, including one confirmed case of E. coli O157 infection. Of the 337 camp attendees, 20 tested positive for E. coli O157 ( Figure 4 ), one of whom tested positive for Cryptosporidium as well. One scout developed HUS and needed dialysis. The attack rate for cub scouts, aged 8–11 years, was 8% (11/145); for scouts, aged 12–16 years, it was 9% (7/81); and for venture scouts, aged 17–25 years, it was 9% (2/22). Dates when symptoms started ranged from May 28 (two cases) to June 3 (one case). Only 25% of the patients had bloody diarrhea. Food and drinking water were ruled out as risk factors, but attendees who did not wash their hands before meals were nearly nine times more likely to be ill with E. coli O157 than those who did. Until the day before the camp, 200 sheep had grazed the grounds, and it was heavily contaminated with feces as well as being waterlogged in places. E. coli O157 was isolated from feces, lying water, soil, Wellington boots, and a temporary climbing frame assembled for the camp; all had the same PFGE type. A detailed study of the grounds after the outbreak concluded that about 50% of the sheep were excreting E. coli O157; ewes, on average, were excreting 8,400/g organisms in their feces. One lamb was excreting more than a million organisms/gram. 24

Epidemic curve, New Deer scout camp 2000 outbreak.
The Godstone Farm outbreak, 2009
The largest farm outbreak in the United Kingdom, and the largest to have been recorded anywhere else, occurred in England in 2009. 5 Godstone Open Farm is in Surrey, to the south of London. In operation for more than 25 years, it had more than 200,000 visitors annually, employing 12 full-time staff with another 12 being taken on part-time at busy periods and 25–30 students being employed at weekends and school holidays. It had two animal petting barns, with the main one housing sheep, goats, pigs, a calf, and a Shetland foal, and the other housing goats, calves, and piglets. All the animals could be touched, and children were encouraged to play with lambs in the main barn. The farm was located in an area which the Surrey and Sussex Health Protection Unit of the Health Protection Agency (HPA) provided public health functions, and Tandridge District Council provided environmental health services. On August 20, the Surrey and Sussex Health Protection Unit was told by the South West London HPU about a child with presumptive E. coli O157 who had visited the farm on August 8, and on that day, another environmental health department informed Tandridge about two other cases. On August 26, the South East London HPU informed the Surrey and Sussex Health Protection Unit about another E. coli O157 case who had visited the farm on August 15, and the next day, another case who had fallen ill on August 24 was reported to the HPU. Tandridge found that this patient had visited the farm on August 21. The farm was advised about extra signage.
The weekend of August 29–31 was a public holiday. On September 1, Tandridge informed the HPU about another case who had fallen ill on August 18. Tandridge and the HPU visited the farm on September 3; the number of confirmed and presumptive cases was now eight. On September 4, the farm voluntarily closed the petting barns. Case numbers continued to rise, but an OTC was not convened (by teleconference) until September 7. It held its second meeting on September 12, when there were 36 confirmed or presumptive cases. The farm closed voluntarily on that day. Media interest became strong on September 13. On September 15, the HPA announced it would set up an independent inquiry to evaluate the outbreak and its management and to consider the regulatory framework and control of risks relating to open farms. The last primary case (infected at the farm) fell ill on September 18, and the last secondary case (infected from contact with a primary case) fell ill on September 22. Transmission on the farm ceased when the petting barns closed. Ninety-three people were infected (91 laboratory-confirmed), with 65 primary cases, 13 secondary cases, and 15 asymptomatic cases. Twenty-seven patients were hospitalized, and 17 developed HUS: ten aged 1–4 years and seven aged 5–9 years. Eight patients needed dialysis, and some have been left with permanent kidney damage. The Pediatric Nephrology Unit at the Evelina Hospital in London serves the whole of South East England. In an average year, it treats five to ten HUS cases. It treated seven cases in September 2009 because of the outbreak. Its facilities were stretched to the maximum. Many E. coli O157 isolates were made on the farm: 25 from ruminants, four from pigs, three from the Shetland pony, one from a rabbit, and two from bird’s nest samples from the main barn and from bark chipping from a bridge and tower in the adventure playground, straw bedding, dust from a metal railing, a pig pen, and a goat pen. All these isolates and the ones from the human cases had variable number tandem repeat profiles indicating they all originated from a common source.
Professor George Griffin chaired the HPA independent investigation. 5 It published interim recommendations in March 2010 and presented a draft final report to the HPA Board on May 26, 2010. It concluded that the OCT was convened exceptionally late and that there was a delay in instituting strict controls at the farm. If contact with ruminants had been stopped on the holiday weekend, or even on Tuesday, September 1, a substantial number of cases could have been prevented. In addition, communication with health care professionals was not discussed at the first OCT (on September 7), and the HPU did not send letters to general practitioners and hospitals until September 10. Pediatric renal units in South East England were not told about the outbreak. There was a lack of public health leadership. The regulatory regime for open farms, including Godstone, was very complex. The farms were inspected by staff from local authority departments of environmental health and trading standards, by Animal Health (an executive arm of the Department for Environment, Food and Rural Affairs), and by the Health and Safety Executive. The HPA, through its local HPU, was tasked to play a lead role in outbreak control, but it had no powers to close an operation. The report made 43 recommendations. Ones identified as particularly important were minimizing visitor contact with animal feces, raising public awareness about this risk, developing an approved code of practice for open farms, getting the regulators to work together, and performing research on rapid diagnosis and the reduction of the carriage of E. coli O157 in animals.
Recent UK outbreaks
An enhanced surveillance system for E. coli O157 and other Shiga toxin-producing E. coli started in England on January 1, 2009. 25 It assisted in the identification of two outbreaks solely on the basis of an increase in a particular phage type in England and Wales.
Phage type 8 caused an average of 22 cases yearly in 2007–2008, 2008–2009, and 2009–2010. During December 2010 and January 2011, 50 cases were confirmed. 26 An increase in cases also occurred in Scotland and Wales. Molecular subtyping showed that many isolates were identical, and an OCT was established. The outbreak lasted until August, with a total of 193 cases in England, 44 in Scotland, and 14 in Wales. Cases occurred over the whole of the United Kingdom, 69% of which were in women. Interviews with 30 sufferers found a statistically significant association with the handling of raw leeks and of potatoes from sacks.
An increase in the number of infections caused by phage type 2 occurred in August 2013: 14 patients fell ill in England, four in Wales, and one in Scotland; 65% were women. Descriptive epidemiology pointed to watercress from a particular retailer and supplier. Microbiological investigations on the watercress and its production sites were negative.
Another outbreak for which the source of infection remains unestablished is one that occurred in Belfast in October 2012. 27 An epidemiological link between cases was eating at Flicks restaurant; 138 cases were confirmed. The OCT report remains unpublished, possibly for legal reasons.
Scotland probably has the highest incidence of E. coli O157 infection in the world. It has maintained this position for many years. North East Scotland usually has the highest incidence within the country. The circumstances surrounding the Rose Lodge outbreak in May 2012 offer a partial explanation. 28 The nursery had 35 infants and children aged 2–5 years registered when the outbreak occurred. It is located in Aboyne, a small town in Aberdeenshire with much animal husbandry in the surrounding area and with many households having private water supplies. The index case was an infant who had probably been infected at home through contact with animal feces either in the surrounding environment or contaminating the domestic private water supply. When tested, the water from this supply was positive for E. coli (not E. coli O157) and coliforms. The infant’s stools were also positive for Cryptosporidium . Three infants and two adults (one was asymptomatic) went on to be infected by person-to-person spread in the nursery, and one adult by person-to-person spread at home. All the isolates had the same relatively rare phage type and indistinguishable pulsed field gel electrophoresis types. One infant developed severe HUS with neurological damage; 20 months after his infection, he is blind and partially deaf and remains in hospital on dialysis with intravenous feeding.
Outbreaks are as relevant to the science of food safety and zoonoses as earthquakes are to seismologists. Outbreak information has led to the quantitation of the infectious dose, the frequency of asymptomatic infection and person-to-person spread, and the time of persistence in the environment, as well as the identification of routes of spread, food vectors, and the age groups most likely to develop HUS. Outbreaks have raised public and political concerns leading to the commissioning of investigations and reports, debates in Parliament, and legislative changes and have influenced the reform of regulatory bodies. A particularly important role in raising public awareness has been played by mothers: Julie Preen set up a fund raising trust in 1999 when her daughter Heather died of HUS (three-case outbreak at Dawlish Warren, source unestablished), and Sharon Mills (mother of Mason Jones) 29 and Tracy Mock (mother of twins who visited Godstone Farm and developed HUS) gave powerful television interviews.
A universal response to tragedy is to say “lessons must be learned”, but the events in South Wales in 2005 repeated those in central Scotland in 1996, so it would be wise to restrict the number of lessons. Two stand out: First, a speedy response is vital. HUS cannot be prevented once an infection has been established. Preventing primary cases by removing the source of infection is obvious, but preventing secondary cases by promulgating hygiene messages to the public is crucially important. 30 Second, there is more to inspecting an operation like a food business than ticking boxes. Personal experience, and even intuition, 31 is very important in detecting ill-intentioned but well-informed operators, 32 such as William Tudor.
Internationally, the United Kingdom has been exceptional in having so many outbreaks linked to butchers (30 recorded between 1995 and 2004 17 ). Unlike the United States, there have been very few cases resulting from the consumption of ground beef (which was the vector in 41% of US foodborne outbreaks between 1982 and 2002 33 ). It is reasonable to suppose that such differences reflect local food preferences and culinary customs.
The enormous German E. coli O104:H4 outbreak in 2011 34 demonstrated that E. coli evolves in real-time. It is likely that many predictions about Shiga toxin-producing E. coli will be wrong, but one certainty is that E. coli O157 in the United Kingdom has not gone away. Laboratories in England, Wales, and Scotland reported 1,039 human isolations in 1995 and 1,029 in 2012. The only good news is that it remains rare relative to other bacterial causes of gastroenteritis; for example, in 2012, the laboratories in these countries 35 reported 71,365 isolations of Campylobacter.
The author reports no conflicts of interest in this work.

Case Study: e Coli Food Poisoning – Taken Out by a Toxic Taco

This case study focuses on the outbreaks of e coli food poisoning similar to those at the Chipotle Mexican Grill which have been in the news recently. With medical algorithms, healthcare providers can quickly distinguish which foodborne illness the patient has and help identify its cause.
Mary is a grandmother with three grandchildren. On Saturday she took them shopping and afterwards they went to their favorite taco restaurant.
On Tuesday Mary felt a little sick when she woke up with some abdominal cramping. Her grandchildren also felt too sick to go to school. Their parents felt fine and the father stayed home to watch the children. Later that day, everyone who had eaten at the taco restaurant developed diarrhea. That evening Mary noticed that her stool was bloody. She developed fever, fatigue, bruising and decreased urine output.
Her daughter took Mary to the Emergency Room early Wednesday morning. Her serum creatinine concentration was high and her platelet count was low. A peripheral blood smear showed numerous schistocytes without platelet clumping. She was admitted to the hospital and started on hemodialysis that afternoon.
The stool culture was positive for Escherichia coli. The isolate was positive for Shiga toxin. A diagnosis of Shiga-toxin producing E. coli (STEC) was made.
Mary was in the hospital for 2 weeks and was discharged home. Recuperation was slow but she gradually recovered without residua.
During her hospitalization, the State Health Department completed an investigation of the restaurant and was unable to find an identifiable source. The restaurant (and its sister locations in the same city) were closed for terminal cleaning and re-opened after no source was found and additional preventive measures were taken.
Shiga-toxin e Coli Poisoning Symptoms & Causes
Shiga-toxin producing Escherichia coli (STEC) can be acquired in several ways, but usually it is acquired from contaminated food. Possible sources include ground beef, poultry, sprouts, cucumbers, lettuce, and other produce. Produce from a single contaminated site may be transported large distances, resulting in multi-state outbreaks.
An infection often presents as a diarrheal disease which must to be distinguished from other food-borne diseases (Salmonella, Shigella, Listeria, Campylobacter, etc). This usually involves stool testing for both bacteria and Shiga toxin.
What makes STEC of concern is that some patients develop hemolytic-uremic syndrome (HUS). This is a much more serious condition which can result in renal and multiple organ failure. HUS overlaps with Thrombotic Thrombocytopenic Purpura (TTP).
HUS or TTP are syndromes that can have multiple causes. Management involves identifying the cause so that specific therapy can be initiated in addition to management of organ failures.
With modern therapy most patients can be successfully treated. Some patients may have prolonged admissions to the intensive care unit. Fatalities can occur in vulnerable patients, patients with delayed care, or fulminant disease. Medical algorithms can help to identify high risk patients who may benefit from more aggressive management.
e coli Prevention
Prevention of STEC infection can be challenging, especially for restaurants. Produce should be carefully cleaned and workers trained in proper food preparation. Meat products should be thoroughly cooked based on internal temperature.
If a case is identified then it is important for food suppliers, restaurants and local health departments to work together to find the source so that further cases can be avoided.
Take-Home Points
- While STEC may be rare, foodborne illness and gastroenteritis is quite common. A powerful algorithm assists physicians in the identification of both common and uncommon pathogens.
- Combining medical training with powerful, evidence-based algorithms can help physicians diagnose, assess, and manage diseases.
The medical algorithms highlighted in this case study are available at The Medical Algorithms Company and also on the a pervita health analytics platform.
About the Authors
Umang Jain is the Health Innovations Fellow at Apervita. He is passionate about medicine, research, and business. He is a fourth year medical student at Northwestern University’s Feinberg School of Medicine and will pursue Emergency Medicine residency. Umang’s scholarly interests include surgical outcomes research, in which he is published in the fields of ENT, orthopedic, plastic, cardiac, and urologic surgery. He has also participated in research in neurodegenerative disease at MIT and Boston University. Umang’s business experience stems from his work at the Institute of Healthcare Improvement (IHI) in Boston, MA. He worked closely with Dr. Donald Berwick, Administrator of Medicare and Medicaid Services (CMS) and Sir Nigel Crisp, the former Chief Executive of UK’s National Health Service, on engaging in evidence-based healthcare improvement interventions on a global scale. Umang was also an intern at Senticare Inc. and Personica, where he evaluated EHRs and in-home health monitoring equipment.
Dr. Chad Rudnick, MD, FAAP is a board-certified pediatrician in Boca Raton, FL. He is the Medical Director of The Medical Algorithms Company. A proponent of incorporating medical technology into his practice, Dr. Rudnick uses telemedicine and medical algorithms from The Medical Algorithms Company in his daily practice to better serve his patients and their families. An accomplished medical writer, he maintains a popular pediatric blog, All Things Pediatric, and has written for numerous online and print publications including KevinMD.com.
John Svirbely, MD is a founder and Chief Medical Officer of The Medical Algorithms Company and the primary author of its medical algorithms. John is a co-founder of the Medical Algorithms Project and has developed its medical content for nearly 20 years. He has a BA degree from the Johns Hopkins University and his MD from the University of Maryland. He is a board-certified pathologist with a fellowship in medical microbiology and biomedical computing at Ohio State University. Currently he is in private practice in Cincinnati, Ohio. He has authored multiple books and articles on medical algorithms.

IMAGES
COMMENTS
Case presentation: We present the case of a patient diagnosed with and exhibiting typical symptoms of CBP. Tests of the prostatic and seminal fluids identified E. coli as the causative pathogen. The patient did not experience favourable long-term treatment outcomes despite repeated antibiotic courses administered over 5 years.
Leafy greens contaminated with Shiga toxin-producing Escherichia coli have continued to cause foodborne illness outbreaks in recent years and present a threat to public health. An important component of foodborne illness outbreak investigations is determining the source of the outbreak vehicle throu …
Background E. coli is an under-recognized cause of bacterial community-acquired pneumonia (CAP). The objective of this study was to describe the epidemiology, risk factors, and outcomes of community-acquired Escherichia coli pneumonia in comparison with other gram-negative and pneumococcal pneumonias.
E. coli's hardiness, versatility, broad palate and ease of handling have made it the most intensively studied and best understood organism on the planet. However, research on E.coli has primarily examined it as a model organism, one that is abstracted ...
By coupling in-depth genomic characterization with a complete sampling of clinical isolates for a full year, this study provides a rare and contemporary survey on the epidemiology and spread of E. coli in a large US healthcare network.
We conducted a matched case-control study and a recipe-based restaurant cohort study, along with environmental, trace-back, and trace-forward investigations, to determine the source of infection.
Risk factors for Escherichia coli O157:H7 infection were investigated in a case-control study at 10 medical centers throughout the United States. Among 73 case-patients and 142 matched controls, exposures in the 7 days before illness associated with E. coli O157:H7 infection in univariate analysis i …
Outbreaks of Escherichia coli O157:H7 infections have involved direct transmission from animals and their environment to humans. We describe an outbreak among visitors to a Pennsylvania dairy and p...
This report describes a large multistate outbreak of Escherichia coli infection linked to a national fast-food chain.
GENERAL INTEREST An Overview of Traceback Investigations and Three Case Studies of Recent Outbreaks of Escherichia coli O157:H7 Infections Linked to Romaine Lettuce Author links open overlay panel Kari Irvin 1 , Stelios Viazis 1 , Angela Fields 1 , Sharon Seelman 1 , Karen Blickenstaff 1 , Ellen Gee 1 , Matthew E. Wise 2 , Katherine E. Marshall 2 , Laura Gieraltowski 2 , Stic Harris 1 Show ...
Background Escherichia coli is a leading cause of bloodstream infections. Developing interventions to reduce E coli infections requires an understanding of the frequency of nosocomial transmission, but the available evidence is scarce. We aimed to detect and characterise transmission of E coli and associated plasmids in a hospital setting.
This case study is largely derived from another study, "An Outbreak of E. coli O157:H7 Associated with Raw Milk," developed in 1994 by Julie R. Crom (Animal and Public Health Inspection Service US Department of Agriculture) and Richard C Dicker, MD, MSc (CDC). This case study was developed in 1998 by Richard C. Dicker.
Shiga toxin-producing E. coli causes clinically important morbidity, particularly in children. Use of optimized diagnostics for identifying STEC and reporting toxin genotypes is essential.
Escherichia coli ( E. coli ), a Gram-negative bacteria, is a member of the family Enterobacteriaceae and is usually a commensal in the gastrointestinal (GI) tract of humans and animals. However, it can acquire mobile genetic elements that confer virulence factors and allow it to become a pathogen that can cause pneumonia, diarrhoea, enteritis ...
Case study and instructor's guide created by: Jeanette K. Stehr-Green, MD NOTE: This case study is based on two real-life outbreak investigations undertaken in Michigan and Virginia in 1997. Some aspects of the original outbreaks and investigations have been altered, however, to assist in meeting the desired teaching objectives and allow completion of the case study in less than 3 hours.
More than forty children in county X contracted what appears to be E. coli 0157. When the state health department announces to the press that there is an E. coli outbreak and that the source is a famous restaurant chain, media interest skyrockets. The public information officer for the state health department asks you to continue fielding ...
Before the COVID-19 pandemic there has been a constant increase in antimicrobial resistance (AMR) of Escherichia coli, the most common cause of urinary tract infections and bloodstream infections. The aim of this study was to investigate the impact of the COVID-19 pandemic on extended-spectrum β-lactamase (ESBL) production in urine and blood E. coli isolates in Finland to improve our ...
Case Study #11: Jack in the Box E. Coli Crisis On January 15, 1993, the Washington State Health Department alerted Robert Nugent, president of Jack in the Box, that the E. coli outbreak they had been informed of two days earlier, was at least partly attributed to hamburgers purchased at Jack in the Box restaurants (Sellnow and Ulmer, 1995).
The antimicrobial activity of silver nanoparticles against E. coli was investigated as a model for Gram-negative bacteria. Bacteriological tests were …
In this case study, we were able to gain valuable knowledge on the efficiency of HPH on E. coli cell disruption, product release and its impact on CQAs. Our results show that HPH is a key unit operation that has to be optimized for each product.
Diarrheagenic Escherichia coli (DEC) are an important cause of acute gastroenteritis in children; however, there is limited information available on the epidemiology, phylogenetics, serotyping, and antibiotic susceptibility of DEC in children in the United ...
Between the months of October and December, 2015, 60 cases of E Coli poisoning linked to Chipotle were reported to the CDC from 14…
E. coli O157 was first isolated during an outbreak in the United Kingdom in July 1983, when stools from three patients tested positive in a cluster of 11 cases of HUS in children from Wolverhampton in the West Midlands. 7 It was a pathogen new to the United Kingdom.
Medical algorithms are highlighted in case study on e coli food poisoning for help with diagnosis, assessment,, treatment, and prevention.