Learn Biology Online
Biology Online is the world’s most comprehensive database of Biology terms and topics. Since 2001 it has been the resource of choice for professors, students, and professionals needing answers to Biology questions. Search over 75,000+ terms, news, insights, discoveries, and trends in Biology below:
- Featured Terms
- Active transport
- Aerobic bacteria
- Alimentary canal
- Allopatric speciation
- Amphipathic
- Anaerobic respiration
- Analogous structures
- Animal cell
- Antagonistic Muscle
- Asexual reproduction
- Assimilation
- Autocrine signaling
- Axon hillock
- Axon terminal
- Balanced diet
- Base-pairing rule
- Binary fission
- Binomial nomenclature
- Biodiversity
- Biological system
- Biotic factor
- Biotic potential
- Biuret test
- Bolus injection
- Bone matrix
- Buck’s traction
- Carbon fixation
- Carrier protein
- Carrying capacity
- Cell morphology
- Cellular respiration
- Chemiosmosis
- Chloroplast
- Chromosomal mutation
- Codominance
- Commensalism
- Complete dominance
- Concentration gradient
- Condyloid joint
- Convergent evolution
- coronavirus COVID-19
- Cross-linking
- Cytokinesis
- Darwinian fitness
- Deciduous forest
- Definitive host
- Dehydration reaction
- Demographic transition
- Dense irregular connective tissue
- Dense regular connective tissue
- Density dependent factor
- Density dependent limiting factor
- Deoxyribonucleic acid
- Desiccation
- Diaphoresis
- Diarthrodial joint
- Dichotomous
- Differentiation
- Disruptive Selection
- Divergent evolution
- Dominant species
- Ecological niche
- Ecosystem diversity
- Endocytosis
- Endomembrane system
- Endosymbiosis
- Endosymbiotic theory
- Energy coupling
- Environment
- Environmental resistance
- Equilibrium
- Eukaryotic cells
- Exotic species
- Facilitated diffusion
- Facultative anaerobe
- Family traits
- Feedback mechanism
- Fermentation
- Fibrinous exudate
- Fibrocartilage
- First-order kinetics
- Fluid mosaic model
- Follicle-stimulating hormone
- Fragmentation
- Frameshift mutation
- Fundamental niche
- Genetic diversity
- Genetic drift
- Genetic material
- Genotypic ratio
- Glycogenolysis
- Golgi apparatus
- Graded potential
- Gram-positive bacilli
- Gram-positive cocci
- Group translocation
- Hermaphrodite
- Heterotroph
- Heterozygous
- Homeostasis
- Homo sapiens sapiens
- Homologous chromosome
- Homologous structures
- Hyaline cartilage
- Hydrophilic
- Hydrophobic
- Hyperosmotic
- Hypersensitivity
- Hypomelanism
- Hypotonic solution
- Hypotonicity
- Immunoglobulin
- Incomplete dominance
- Incubation period
- Independent variable
- Induced fit model
- Inoculation
- Inorganic compound
- Intercalary meristem
- Intermediate host
- Interspecific competition
- Interstitial fluid
- Intracellular digestion
- Involuntary muscle
- Iodine test
- Jaw jerk reflex
- Joint capsule
- Joints of pelvic girdle
- Jugulo-omohyoid node
- Juncturae cinguli membri superioris
- Kempner diet
- Keratinocyte
- Kidney calices
- Kilocalorie
- Kinetochore
- Kingdom Animalia
- Kleptoparasitism
- Krebs cycle
- Law of Dominance
- Law of Independent Assortment
- Law of Segregation
- Lethal mutation
- Leukocytosis
- Light energy
- Limiting factor
- Longitudinal section
- Loose associations
- Loose connective tissue
- Memory cell
- Micromolecule
- Missense mutation
- Mitochondrion
- Monosaccharide
- Multiple alleles
- Muscular system
- Mutualistic symbiosis
- Naked virus
- Natural selection
- Negative feedback
- Nerve impulse
- Net photosynthesis
- Net primary productivity
- Neutral mutation
- Non Random Mating
- Non-living thing
- Nondisjunction
- Nuclear envelope
- Null hypothesis
- Nulliparity
- Obligate aerobe
- Obligate anaerobe
- Obligate parasite
- Observation
- Okazaki fragment
- Oligosaccharide
- Open circulatory system
- Opportunistic pathogen
- Opsonization
- Organic compound
- Organic matter
- Organic molecule
- Organization
- Osmotic Potential
- Osseous tissue
- Parthenogenesis
- Passive transport
- Phagocytosis
- Phenotypic ratio
- Phosphodiester bond
- Physiological adaptation
- Pinocytosis
- Pioneer species
- Polygenic inheritance
- Polypeptide
- Positive feedback
- Primary succession
- Protein synthesis
- Punnett Square
- Quadrangular cartilage
- Quadrantanopia
- Quantitative analysis
- Quantitative trait
- Quaternary structure
- Quellung phenomenon
- Quinacrine banding
- Radial symmetry
- Realized niche
- Redox reaction
- Reducing sugar
- Relative fitness
- Replication
- Reproduction
- Reservoir host
- Respiration rate
- Resting potential
- Reticular connective tissue
- Ribonucleic acid
- Secondary consumer
- Selectively-permeable membrane
- Semilunar valve
- Sex-linked trait
- Sexual reproduction
- Simple diffusion
- Simple squamous epithelium
- Single stranded DNA virus
- Skeletal system
- Smooth endoplasmic reticulum
- Smooth muscle
- Sympatric speciation
- Synergistic effect
- Tap water enema
- Telophase II
- Thalassophobia
- Thermophile
- Thin filament
- Transcription (biology)
- Translation
- Triploblastic
- Trophic level
- True breeding
- Turgor pressure
- Ultradian rhythm
- Unconditioned stimulus
- Undergrowth
- Undulating membrane
- Unicellular
- Unipennate muscle
- Unipotent cell
- Universal solvent
- Unmyelinated neuron
- Unsaturated fatty acid
- Unsustainability
- Urinary system
- Uterine cycle
- Uterine involution
- Vascular plants
- Vegetative reproduction
- Ventilation
- Vertical gene transfer
- Voice quality
- Voluntary muscle
- Wagners syndrome
- Waste management
- Wasted ventilation
- Water potential
- Water table
- White adipose tissue
- White blood cell
- White line of toldt
- Wuchereria bancrofti
- X chromosome
- X-linked inheritance
- Xanthophyceae
- Xanthophyll
- Xanthophyte
- Xerophthalmia
- Xxy syndrome
- Xylem fiber
- Xylem parenchyma
- Xylem vessel
- Y chromatin
- Y chromosome
- Y-linked gene
- Y-linked inheritance
- Yellow fever virus
- Young-helmholtz theory of colour vision
- Zone of inhibition
- Zooflagellate
- Zoogeography
- Zoomastigophora
- Zooplankton
- Zymogen granules

Featured Forum Topics
Explore our Forums and discover the living world. Feel free to post your thoughts and questions, and get answers from our esteemed online community.
- Where does protein synthesis take place
- What is phylum Chordata?
- What does mRNA do in protein synthesis?
- Sympatric vs allopatric speciation
- Asexual and sexual reproduction- differences!
- Advantages and disadvantages of asexual reproduction
- Difference between homologous chromosomes and sister chromatids
- Smooth muscle vs dense regular connective tissue
- What is the key to the recognition of codominance?
- Incomplete dominance vs codominance
Biology Articles
Keep up with the latest trends and new insights in the field of Biology.

Bondok’s Classification of the Thalamic Nuclei
Anatomists usually classify the thalamic nuclei according to their relation to the internal medullary lamina i..

Mendel’s laws: definitions
Genetics – a branch of biology that deals with the study of heredity and variations Variations – d..

Gorillas in our Midst – what it means
Have you tried gorilla on a basketball court perception test? Do you know what it implicates? Find out in this..
Public Engagement: Rising Expectations in the Digital Age
Gone are the simpler days where scientists could isolate and focus solely on their work, engaging exclusively ..
Biology Tutorials
Working on a research paper, or preparing for an exam. Our Biology Tutorials will set you up for success.

Freshwater Ecology
Freshwater ecology focuses on the relations of aquatic organisms to their freshwater habitats. There are two forms of co..

Lights’ Effect on Growth
This tutorial elaborates on the effect of light on plant growth. It describes how different plants require different amo..

Plant Water Regulation
Plants need to regulate water in order to stay upright and structurally stable. Find out the different evolutionary adap..

The Evolutionary Development of Multicellular Organisms
Multicellular organisms evolved. The first ones were likely in the form of sponges. Multicellularity led to the evolutio..
- Skip to primary navigation
- Skip to main content
- Skip to footer
Bringing the World's Best Biology to You

iBiology and iBiology Courses are part of the Science Communication Lab (SCL). Our mission remains the same, to connect people to science. However, our focus has shifted to producing and evaluating cinematic films for education and the public, which you can find on the Science Communication Lab website . Please visit us there for new content and videos!
Search our library of 600+ biology videos including lectures, talks, and animations:
Free biology lecture videos.
Search our library of hundreds of in-depth biology lecture videos presented by the scientists behind the discoveries.
Free Online Courses for Scientists
We offer a bevy of free professional development and research skills courses for early career scientists and trainees.
Free Educator Resources
We provide free educator resources for our content, from learning objectives to our popular monthly newsletter.
Best of iBiology
We’ve produced hundreds of free videos since 2006. Here are some of our top videos and recommended lectures for education, learning techniques, and just for fun!
Learn from Scientists
Dive into the science behind world-changing discoveries , presented by the scientists themselves.
CRISPR-Cas9
Jennifer Doudna shares how studying the way bacteria fight viral infection turned into a genomic engineering technology that has transformed molecular biology research.

Finding Tiktaalik
Neil Shubin unveils the careful planning that was key to the discovery of Tiktaalik roseae, the 375 million year old fossil link between fish and land animals.

Molecular Motor Proteins
Lasker Award winner Ron Vale gives an overview of the molecular motor proteins that power much of the movement performed by living organisms.
Viruses & Phage Therapy
Paul Turner describes the fundamental biology of viruses, and provides an introduction to phage therapy, and how it can be improved by applying evolution thinking.

Archaea and the Tree of Life
Dipti Nayak explains how the mysterious microbes known as archaea are helping scientists rewrite the tree of life as well as her research on methanogenic archaea.

Immunology & B Cell Development
Shiv Pillai provides a historical overview of today’s model on how the immune system works and outlines the underpinnings of B cell development.
- Free Online Courses
From designing experiments to effective science communication to professional development, we offer a bevy of self-paced online courses to help you plan your career and build critical research skills. Sign up today for our free platform and join a community of thousands of early career scientists, students and educators in biology!

Microscopy & Other Techniques
Learn the theory and application of microscopy and other techniques in research .

- Learn Microscopy
Everything you need to know about microscopy from basic to advanced techniques with lectures, labs, and assessments. We also offer a short 14 lecture series for those new to microscopy.

- Next Generation Sequencing
Next generation sequencing allows DNA samples to be sequenced quickly and affordably. Learn from Eric Chow how next gen sequencing works and get tips on preparing and running your samples.

- Bioimage Analysis
This series follows the life cycle of an image data set, from acquisition to analysis and provides important concepts, best practices, and examples that will come in handy for scientists designing their own experiments.
Recommended Series & Playlists
Explore further with popular series and curated playlists.

Share Your Research
Early career scientists share their research stories after receiving intensive science communication training from iBiology.

- Famous Discoveries
Join some of the world’s most well-known scientists as they share the details and science behind their major discoveries.

- All Playlists
From talks on genomics to talks with Nobel Prize winners to white-board animations, find all of our curated playlists here!
Want more educator resources and science films?

iBiology is part of the Science Communication Lab .
Find a video.
- Search Videos
- All Speakers
- YouTube Channel
- Other Languages
- Biochemistry
- Bioengineering
- Cell Biology
- CRISPR-Cas Technology
- Development & Stem Cells
- Genetics & Gene Regulation
- Human Disease
- Microbiology
- Neuroscience
- Plant Biology
- Science and Society
For Educators
- New! SCL Films for Education
- New! SCL Educator Newsletter
- iBiology Educator Resources
- iBiology Flipped Courses
- iBiology Courses Resources
- Recommended Playlists
- View All Educator Resources
Educators Recommend
- Cells of the Immune System
- Discovering Cell Cycle Regulators
- Genome Engineering with CRISPR-Cas9
- Hardy-Weinberg Equilibrium
- Semi-Conservative Replication of DNA
- Toll-Like Receptors in Adaptive Immunity
- Intro to Microscopy
- Intro to Flow Cytometry
- View All Techniques
Free Courses
- Launch iBiology Courses
- Cell Biology Flipped Course
- Engineering Life Flipped Course
- Evolution Flipped Course
- Immunology Flipped Course
Career Development
- Career Development Courses
- Career Development Videos
- Career Exploration Videos
- Biomedical Resources
- Biomedical Workforce
- View All Career Development
Our Favorites
- Short Films & Stories
- Feature Documentaries
- Improving Diversity in Science
- Share Your Research Series
Connect with Us
- iBiology’s Mission
- Contact iBiology
- Video Usage & Licensing
- Sign Up for Our Newsletter
- Subscribe to YouTube Channel
- Go to the Science Communication Lab

- Drug resistance
- Genetic Engineering
- Microbiology
- Paleontology
- Sex & Gender

‘Frog Saunas’ Could Protect Species from Devastating Fungal Disease
A low-tech immune boost may help some species of frogs survive a brutal fungal disease that’s already ended 90 species
Meghan Bartels

Marijuana’s New Legal Status Must Spark More Research
While most Americans enjoy access to marijuana, barriers to research and knowledge expansion persist—with dangerous consequences. Unlimited research access is needed
Joerg Leheste
These Hormones Drive Bloodlust in Mosquitoes
Mosquitoes carry a pair of hormones, one of which drives bloodlust while the other signals satiation, scientists say
Gemma Conroy, Nature magazine

Pupil Dilation Reveals Better Working Memory
People whose eyes dilated more performed better on tests of working memory
Kate Graham-Shaw

A Retracted Stem Cell Study Reveals Science’s Shortcomings
The withdrawal after 22 years of a controversial stem cell paper highlights how perverse incentives can distort scientific progress
Peter Aldhous

How Delicate Comb Jellies Withstand Crushing Depths—But Melt Away on Land
Scientists finally know how a gelatinous deep-sea creature keeps its cells from paralysis under pressure
Elizabeth Anne Brown

Highly Invasive Spotted Lanternflies May Have a Surprising Weakness: Vibrations
Spotted lanternflies are sometimes drawn to power line vibrations—and scientists are taking notice
Claire Marie Porter

To Follow the Real Early Human Diet, Eat Everything
Nutrition influencers claim we should eat meat-heavy diets like our ancestors did. But our ancestors didn’t actually eat that way
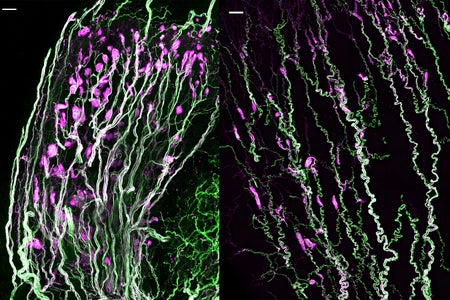
Sensory Secrets of Penis and Clitoris Unlocked after More Than 150 Years
Mysterious nerve structures called Krause corpuscles respond to specific low-frequency vibrations, scientists finally confirm
Sara Reardon, Nature magazine

Great Tits Show How Animals Can Thrive in Cities
One of Europe’s most common birds, the great tit, show an amazing adaptability to human-made habitats. There seem no limits for this species when it comes inventing new ways of acquiring food from people
Anders Brodin

These Bloodsucking Leeches Jump like Striking Cobras
Scientists observed leeches jumping like striking snakes, resolving long-standing debate
Gennaro Tomma

Out of Sight, ‘Dark Fungi’ Run the World from the Shadows
The land, water and air around us are chock-full of DNA from fungi that scientists can’t identify
Cody Cottier
Empowering Educators. Inspiring Students.
Real science, real stories, and real data to engage students in exploring the living world.

Unlock the full power of BioInteractive!
Create a free account to access new features and join our Online Community exclusively for verified educators.
- Connect with educators
- Access teaching tools
- Explore the Educator Resource Library
- Create custom resource collections
- Staff Picks
- Most Viewed

Setting Up Your Data for Success
This highly customizable activity guides students through organizing and documenting spreadsheet data during a class investigation.

Evolution in Action: Data Analysis
In this activity, students analyze and graph data taken from a population of Galápagos finches before and after a major drought.

Beaks As Tools: Selective Advantage in Changing Environments
In this activity, students collect and analyze data from a hands-on model to discover why even slight variations in beak size can impact a bird’s ability to obtain food and survive.

Interactive Assessment for The Beak of the Finch
A number of questions are embedded within the short film The Origin of Species: The Beak of the Finch , which explores four decades of research on the evolution of the Galápagos finches.

Sampling and Normal Distribution
This interactive simulation allows students to graph and analyze sample distributions taken from a normally distributed population.

Sorting Finch Species
This interactive module allows students to explore concepts related to speciation by identifying which birds belong to one of two finch species.

What Is Life?
In this activity, students consider the challenges of defining life by sorting cards of living and nonliving things and comparing their characteristics.

Smallpox and the Immune System
This activity uses two short historically based stories about smallpox, told through comics, to motivate learning about the immune response, immune cells, and vaccines.

The Genetics of Tusklessness in Elephants
This video follows scientists working in Gorongosa National Park as they try to determine the genes responsible for tusk development in elephants.

Trees on the Serengeti
This activity explores images of tree growth in the Serengeti over time, which serve as a phenomenon for learning about and modeling species interactions in ecosystems.

Digestive System Card Sort
In this card activity, students investigate the functions of major digestive organs.

Sickle Cell: Natural Selection in Humans
This film explores the evolutionary connection between an infectious disease, malaria, and a genetic condition, sickle cell disease.

The Eukaryotic Cell Cycle and Cancer
This interactive module explores the phases, checkpoints, and protein regulators of the cell cycle. The module also shows how mutations in genes that encode cell cycle regulators can lead to the development of cancer.

Lizard Evolution Virtual Lab
This interactive, modular lab explores the evolution of the anole lizards in the Caribbean through data collection and analysis.

Virus Explorer
This interactive module explores the diversity of viruses based on structure, genome type, host range, transmission mechanism, replication cycles, and vaccine availability.

The Biology of Skin Color
This film explores the hypothesis that different tones of skin color in humans arose as adaptations to the intensity of ultraviolet radiation in different parts of the world.

Photosynthesis
This multipart animation series explores the process of photosynthesis and the structures that carry it out.

EarthViewer
This interactive module allows students to explore the science of Earth's deep history, from its formation 4.5 billion years ago to modern times.
Educator Voices
Hear how experienced science educators are using BioInteractive resources with their students. Discover implementation ideas, lesson sequences, resource modifications, quick tips, and more in this collection of videos and in-depth articles.

Crash Course Biology in Collaboration with BioInteractive
HHMI BioInteractive is a proud collaborator on a new project that reimagines the original Crash Course Biology series. In this article, science communicator Hank Green discusses how this collaboration will deliver value for both students and teachers.

Getting Students Asking Scientific Questions Using BioInteractive Resources
Asking scientific questions is a foundational skill that takes instructional support for students to develop. In this article, Bernice O'Brien outlines how she uses BioInteractive resources to get her students to formulate and refine scientific questions.

Developing the Model Builder Web Tool
To develop our Model Builder web tool, BioInteractive worked with Jon Darkow, an educator with extensive experience in using models with his students. In this video and Q&A, Jon discusses the Model Builder features he’s most excited to share with other educators.

Evidence-Informed Tips for Using the Interactive Video Builder Tool
Our new Interactive Video Builder tool lets educators embed their own questions into our videos. In this Educator Voices article, Annie Prud’homme-Généreux details research-based strategies for designing effective interactive videos.

A Virtual Animal Behavior Research Project for an Introductory Biology Course
Professor Melissa Haswell details a multiweek virtual model to develop basic scientific knowledge and skills using BioInteractive resources that culminates in an eight-week-long animal behavior research project.

Modeling Cellular Respiration for Relevance and Reasoning Using BioInteractive Resources
In this Educator Voices article, professor John Moore describes a "backwards" approach to teaching energy use in cells that traces the process from ATP in use back through glycolysis.

Teaching About the Cell Cycle, Immune System, and Cancer Using BioInteractive Resources
Getting students engaged in learning about the cell cycle can be difficult. In this Educator Voices article, educator Kathy Van Hoeck describes how she uses cancer as an anchoring phenomenon to spark student interest.
Teaching Support

Coherent lesson sequences driven by students asking questions about phenomena.

Resource Playlists
Ordered sequences of BioInteractive resources for teaching a course, unit, or lesson.

Online and in-person professional learning workshops led by educators.

Science News
Articles that connect current events to BioInteractive resources.
Discover tools to help plan lessons and opportunities to support professional learning.
Announcements
Keep up with the latest from BioInteractive!

Developing Educators as Leaders
The HHMI BioInteractive Ambassador Academy is a three-year professional development experience designed to promote and support evidence-based teaching practices that incorporate our approach and values as well as our free and accessible classroom resources.

New Online Professional Development Workshops
BioInteractive is offering free workshops for high school and undergraduate life and environmental science educators. All workshops are online, facilitated by Ambassadors, and include opportunities to interact with our resources and learn from other educators.

HHMI Expands Commitment to Diversity, Equity, and Inclusion
HHMI is investing in increasing racial, ethnic, and gender diversity in academic science to create environments in which everyone can thrive.

Racial Inequities and Our Work
BioInteractive is committed to providing equitable learning opportunities to educators and students. We know we have a lot of work to do to address racial inequities in science teaching. The following statement reflects our current and specific actions.

Introducing a new BioInteractive experience
- COVID-19 Tracker
- Biochemistry
- Anatomy & Physiology
- Microbiology
- Neuroscience
- Animal Kingdom
- NGSS High School
- Latest News
- Editors’ Picks
- Weekly Digest
- Quotes about Biology

Biology Dictionary
Biology is the study of living things. It is broken down into many fields, reflecting the complexity of life from the atoms and molecules of biochemistry to the interactions of millions of organisms in ecology. This biology dictionary is here to help you learn about all sorts of biology terms, principles, and life forms. Search by individual topic using the alphabetized menu below, or search by field of study using the menu on the left.
Trending Biology Topics
Acetic acid.
- Active Transport
Adenosine Triphosphate (ATP)
- Adrenal Gland
- Aldosterone
- Amino Acids
Animal Cell
- Biodiversity
- Carbon Cycle
- Cell Membrane
- Cellular Respiration
- Chlorophyll
- Chloroplast
- Circulatory System
- Commensalism
- Condensation
- Cytokinesis
- Digestive System
- Endocrine System
- Endoplasmic Reticulum
- Facilitated Diffusion
- Formaldehyde
- Golgi Apparatus
- Hermaphrodite
- Heterotroph
- Heterozygous
Homeostasis
Horticulture, hydrochloric acid.
- Integumentary System
- Krebs Cycle
- Lymphatic System
- Metamorphosis
- Mitochondria
- Muscular System
- Natural Selection
- Nervous System
- Nitrogen Cycle
- Nucleic Acid Elements and Monomer
- Nucleic Acid Types and Structure
- Phagocytosis
- Phospholipid
- Photosynthesis
- Pineal Gland
Pituitary Gland
- Punnett Square
- Respiratory System
- Rigor Mortis
- Scientific Method
- Skeletal System
- Sulfuric Acid
- Sympathetic Nervous System
- Three Parts of Cell Theory
- Thyroid Gland
- Transcription
- Translation
- Transpiration
- Water Cycle
Privacy Policy
Terms of service, scholarship, latest posts, white blood cell, t cell immunity, satellite cells, embryonic stem cells, popular topics.

The Biology Corner
Biology Teaching Resources

The Biology Corner – Resources for Teachers

Biology 1 & 1A Biology 2 & 2A AP Biology Anatomy 1 & 2 Intro Biology

CK 12 Reading Guides OpenStax Reading Guides Miller Levine (Dragonfly)

Animals of North America Coloring Book

Dissections
Fetal Pig Frog / Bullfrog Squid Cat Rat Earthworm Heart / Brain
Assessments
Kahoot Quizziz Quizlet Practice Quizzes
Teachers Pay Teachers

Support Biology Corner
All student resources are available for free.
To support this site, purchase answer keys at TpT or DONATE via PayPal

I have taught biology for 30+ years and have uploaded and shared resources throughout my career. You are free to use and edit any of the documents. Tests and answer keys can be purchased from TpT.

Dissection Photos
Biology Clipart
Branches of Biology
- Importance of Biology
- Domain Archaea
- Domain Eukarya
Biological Organization
- Biological Species Concept
Biological Weathering
- Cellular Organization
- Cellular Respiration
- Types of Plants
- Plant Cells Vs. Animal Cells
- Prokaryotic Cells Vs. Eukaryotic Cells
- Amphibians Vs. Reptiles
- Anatomy Vs. Physiology
- Diffusion vs. Osmosis
- Mitosis Vs. Meiosis
- Chromosome Vs. Chromatid
History of Biology
- Biology News
Bio Explorer

The Top 25 Bicolor Flowers: Nature’s Two-Toned Wonders
Top 18 amazon rainforest plants, top 26 best hawaiian flowers, 25 must-see colorful orchids, what do peacocks eat, welcome to bioexplorer.

B ioExplorer shares breaking science news and articles on a variety of topics from the leading universities and research institutions around the globe. It is your ultimate guide to Biological Web Resources ranging from DNA to Plants, Animals, and everything in-between.
This site was originally developed for the researchers involved in biological studies, but we want to make the science more enjoyable to even a common man too. Stay tuned for the exciting gardening, book reviews, science news articles to be updated here regularly. Read More
Biology News By Year

Top 15 Anatomy And Physiology News of 2022
Top Immunology News of 2022

Top 15 Genetics News of 2022

Top 15 Evolutionary Biology News of 2022

Top 15 Biochemistry News of 2022

Top 15 Ecology News of 2022

Top 15 Cell Biology News of 2022

Top 15 Biotechnology News of 2022

Top 15 Botany News of 2022

Top 15 Microbiology News of 2022

Top 15 Anatomy News of 2021

Top 15 Immunology News of 2021

Top 15 Genetics News of 2021

Top 15 Evolutionary Biology News of 2021

Top 15 Biochemistry News of 2021

Top 15 Ecology News of 2021

Top 15 Cell Biology News of 2021

Top Biotechnology News of 2021

Top 15 Botany News of 2021

Top 15 Microbiology News of 2021

Top 10 Anatomy/Physiology News in 2020

Top 10 Immunology News of 2020

Top 10 Genetics News of 2020

Top 10 Evolutionary Biology News of 2020

Top 10 Biochemistry News of 2020 – A Round-Up

Top 10 Ecology News of 2020

Top Cell Biology News of 2020 – A Round Up

Top 10 Biotechnology News of 2020

Top 10 Botany News in 2020

Top 10 Microbiology News of 2020

Top 10 Discoveries in Immunology 2019

Top 10 Discoveries in Ecology 2019

Latest Discoveries In Microbiology 2019

Top 10 BEST Genetics Discoveries of 2019

Top 2019 Discoveries in Evolution

Top 10 Cell Biology Discoveries in 2019

Top 10 BEST Botany Discoveries in 2019

Top 10 Biotechnology Discoveries in 2019

Top 15 Biochemistry Discoveries of 2019

Top 15 Anatomy & Physiology News In 2019

Top 26 Anatomy & Physiology News in 2018

Top 15 Biochemistry Discoveries in 2018

Top 25 Most Recent Genetic Discoveries in 2018

15 Wonderful Biotechnology Inventions In 2018

Top 15 Discoveries in Cell Biology for 2018

15 Latest Inventions in Botany For 2018

Top 15 Evolutionary Biology News in 2018

15 Leading Ecology News of 2018

Top 15 Latest Microbiology & Virology Discoveries in 2018
Top 15 Immunology News in 2018
Top 15 Anatomy & Physiology News In Innovations & Breakthroughs For...

Top 14 Biochemistry News, Innovations & Breakthroughs In 2017

Top 12 Botany News For 2017
Top 12 Cell Biology News in 2017

Top 10 Ecology News in 2017

Top 12 Genetics News In 2017

Top 12 Immunology News In 2017

Top 11 Microbiology News In 2017

Top 10 Biotechnology News In 2017

Top 10 Evolution News In 2017

How To Collect Rainwater For Gardening?

What Vegetables Are Good For Vertical Gardening?

Top 10 Best Gardening Tools For Dad

Best Indoor Plant Lighting For Growing

10 Best Low Maintenance Plants For Indoors

6 Best Indoor Vegetable Garden Systems Reviews

Exploring the Top 50+ Most Exquisite Purple Flowers in the World

25 Most Known Exotic Flowers

25 Most Famous & Dangerous Bug Eating Plants

Types of Flowers

Bell-Shaped Flowers

Cone-Shaped Flowers

Star-Shaped Flowers

Thread-Shaped Flowers

Top 100 BEST Fragrant Flowers

Top Short-lived Flowers

The Ultimate Guide to 25+ Spectacular Desert Flowers & Their Unique Adaptations

Flowers That Start with A

Flowers Starting with B
Recently on bioexplorer.

World’s Top 15 Poisonous Caterpillars

35 Most Colorful Birds In The World!
Best recommended universities / courses.

Top 15 USA Biology Scholarships For Aspiring Biology Students

Top 10 BEST Colleges For Nutrition and Dietetics

Top 10 BEST Colleges For ObGyn

Best Colleges For Environmental Engineering

Top 10 Best Colleges For Biomedical Engineering

Top 10 Best Universities For Veterinary Medicine

Top 10 BEST Nursing Schools in California

Top 14 BEST Colleges For Dentistry

Top 10 BEST Colleges For Biochemistry

Top 10 MCAT Prep Books – An Ultimate Guide

How To Become A Sports Medicine Physician?

Top 15 Wildlife Biology Degree Programs In The USA

Top 15 Online Microbiology Courses For US Students

Top 10 Best Neurosurgery Schools in America
Browse categories.

Explore the Animal kingdom and different types of animals ( monkeys , apes , crocodiles , birds & more) here.

Learn all about bacteria, diseases caused by bacteria and more here.

Biology Apps
Best & Free biology apps for students and teachers.

Biological Weathering is a natural phenomenon that occurs on rocks due to living organisms.

Learn about biologists around the world who made their impressions in the study of biology.

Bioethics can rightly be described as a combination of two subjects – Biology and Philosophy.

Identify different branches of biology here.

Differences Between
Explore the difference between various two biological concepts here.

Biology-themed Movies
Explore the best biology-themed movies and documentaries

Fathers of Biology
Learn the pioneers (aka Fathers of Biology) in different sub-branches of biology and their scientific contributions.

Biological Journals
Find Biological Journal collections from Anatomy to Zoology.

Biological Libraries
Biology library and literature resources from around the globe.

Biology Online Tools
Online tools related to biological studies especially on Bioinformatics and other fields.

Biological Search Engines
Find a collection of search engines and directories around biological studies.

Biology Softwares
A collection of Software programs (free & paid) useful in biological studies.

Types of Doctors
A list of common types of doctors and their specializations in different areas in the field of health care profession categorized alphabetically.

Gardening 101
Explore gardening-related articles, tips, reviews and recommendations.

Discover the different types of plants , types of flowers , types of trees and all plant-related topics here.

Biological Magnification
Explore the concept of biological magnification here.

Biology Books
Explore the best-rated biology books arranged by sub-disciplines.

Learn 10 levels of biological organization here.

Biology Yearly News
Explore yearly biology news round-ups categoried by various sub-disciplines of biology.

Explore the world of dinosaurs and famous paleontologists here.

Biological Species Concepts
Define species using the Biological Species Concept.

Biological Discoveries
Explore top 25 biological discoveries of all times here.

Pros and Cons
Explore the pros and cons of a biological term or concept here.

Biological Databases
Browse major sequence databases, protein databases, protein domains and RNA databases.

Biology Scholarships
Browse top scholarships for biology majors in the USA.

Biology Questions
Explore curiosity building biology related questions and answers here.

Biology Jobs
Explore top US cities for biology-related job opportunities.

Go back in time machine to learn about advent of life on earth, right from the ancient times.

Genetic Determinism / Biological Determinism
Explore what is genetic determinism, its history, concepts and types here.

Biological Research Institutes
Research Institutes: Biology, Biochemistry, Molecular biology, Zoology and much more.

Methods & Protocols
All protocols in biology are divided into categories such Biochemistry, Cell Biology and more.
Recent Posts

Top Spectacular Rainforests of The World

Top 21 Facts About Blue Morpho Butterfly


What Animals Live In Rainforests?

America’s 15 Must-Visit Botanical Gardens: Discover Them Today!

43 Blue Flowers Unveiled: Why These Beauties Have Captured Everyone’s Attention?

Top 18 BEST Tundra Animal Adaptations
Marine Biology 101: Ocean Life Explained

25 Reasons That Emphasizes The Importance of Biology

Top 27 Biology-themed Movies

Biology Boomtowns: 10 Best US Cities for Job Opportunities

Uncovering the Fathers of Biology: The Geniuses Who Unveiled Life’s Secrets

25 Mind-Blowing Biology Breakthroughs That Shaped Our World!

Discover 162+ Doctor Specialties You Need To Know!

40 Different Types of Birds

334 Types of Monkeys
Biology history.

History of Anatomy

History of Biochemistry

History of Biotechnology

History of Botany
History of Cell Biology

History of Ecology

Complete History of Evolution

History of Genetics

History of Immunology

History of Microbiology
- Privacy Policy

The Biology Notes
Top and Best Biology Websites or Blogs of 2023 for Study Notes
World Ranking and estimated montly website visitors is based on the data obtained from similarweb . We update the website/blog ranking only once in a three month . If your website/blog is not listed below or we missed it, you can email your website/blog URL to microbenotes(at)gmail(dot)com and we will update it on this page.

These are the best biology websites or blogs that provide educational study notes and information to students and teachers. We have only included here individual blogs created by individual persons. We have not included websites as a company or professionals that provide study notes like Khan Academy, Spark Notes, BYJU’s, Toppr, ThoughtCo, Cliffsnotes, Vedantu, etc.
Table: Top biology blogs and websites with a similarweb estimate of monthly visitors and world ranking. (As of November 2022)
| 1 | biologydictionary.net | 1,200,000 |
| 2 | biologyonline.com | 966,600 |
| 3 | microbenotes.com | 951,300 |
| 4 | bioninja.com.au | 942,700 |
| 5 | biologydiscussion.com | 742,700 |
| 6 | microbiologyinfo.com | 595,100 |
| 7 | microbeonline.com | 454,300 |
| 8 | biologycorner.com | 273,300 |
| 9 | microbiologynote.com | 205,800 |
| 10 | onlinebiologynotes.com | 188,400 |
| 11 | bioexplorer.net | 187,000 |
| 12 | thebiologynotes.com | 178,800 |
| 13 | microscopemaster.com | 142,200 |
| 14 | alevelbiology.co.uk | 138,200 |
| 15 | easybiologyclass.com | 121,300 |
| 16 | biologyreader.com | 104,800 |
| 17 | notesonzoology.com | 99,800 |
| 18 | microbiologynotes.org | 88,500 |
| 19 | biologyexams4u.com | 85,200 |
| 20 | laboratoryinfo.com | 82,900 |
| 21 | biodifferences.com | 65,900 |
| 22 | paramedicsworld.com | 63,000 |
| 23 | microbiologyinpictures.com | 56,900 |
| 24 | biologyjunction.com | 52,400 |
| 25 | learn-biology.com | 49,800 |
| 26 | microbiologynutsandbolts.co.uk | 46,200 |
| 27 | onlinesciencenotes.com | 41,300 |
| 28 | biology4kids.com | 39,600 |
| 29 | biologyeducare.com | 39,600 |
| 30 | themedicalbiochemistrypage.org | 38,300 |
| 31 | rsscience.com | 37,100 |
| 32 | biotechnologynotes.com | 35,000 |
| 33 | bacteriainphotos.com | 33,900 |
| 34 | biology-pages.info | 31,700 |
| 35 | biomadam.com | 28,400 |
| 36 | biologyease.com | 22,700 |
| 37 | microbiologynotes.com | 21,800 |
| 38 | overallscience.com | 18,000 |
| 39 | biosciencenotes.com | 16,800 |
| 40 | biologyreference.com | 15,700 |
| 41 | thebiologyprimer.com | 13,000 |
| 42 | thesciencenotes.com | 11,100 |
| 43 | microbe.net | 8,700 |
| 44 | awkwardbotany.com | 8,000 |
| 45 | wildlifebiology.org | 7,300 |
| 61 | 24hoursofbiology.com | <5000 |
| 61 | askmicrobiology.com | <5000 |
| 61 | biologybrain.com | <5000 |
| 61 | biologyguide.app | <5000 |
| 61 | biologyideas.com | <5000 |
| 61 | biologynotes.site | <5000 |
| 61 | biopharmanotes.com | <5000 |
| 61 | concisebiology.com | <5000 |
| 61 | generalmicroscience.com | <5000 |
| 61 | learnbiology.net | <5000 |
| 61 | microbes.info | <5000 |
| 61 | microbesinfo.com | <5000 |
| 61 | microbialfacts.com | <5000 |
| 61 | microbialmenagerie.com | <5000 |
| 61 | microbiologymatters.com | <5000 |
| 61 | sciencevivid.com | <5000 |
Here are more descriptions of the biology-related blogs with a similarweb estimate of monthly visitors ranking from 1 to 61.
Table of Contents
1. biologydictionary.net
This website’s mission is to be the definitive source of meaningful and informative explanations of biological concepts, to anyone who wants to learn. This website covers all the basics of biology with concept articles, comparison articles, and basic definitions of everything biology-related! They offer quizzes by topic or category and are currently working on a number of other study guides and aides. They are working on video tutorials, helpful infographics of each topic, study guides for students, lesson plans for teachers, and a number of other useful products.
Website URL: https://biologydictionary.net/
2. biologyonline.com
Biology Online is the home for everyone who is interested in studying and becoming more familiar with Biology, whether you are a Student, Educator, Scientist, Life Science professional, or anyone else who shares a passion for Biology. Created in 2001, the site is the world’s most comprehensive database of Biology terms, tutorials, and articles with over 33 million visitors using the site on an annual basis. The site aims to educate and promote awareness of all things regarding Biology, offering free and easy access to foundational information in the Biological Sciences.
Website URL: https://www.biologyonline.com/
3. microbenotes.com
Microbe Notes is an educational niche blog/website related to microbiology (bacteriology, virology, parasitology, mycology, immunology, molecular biology, biochemistry, etc.) and different branches of biology with an aim to provide study notes to undergraduate and graduate students. This website is also useful for A-level biology, AP biology, IB biology, and other university-level biology and microbiology courses (B.Sc, M.Sc., M.Phil., and Ph.D.).
Website URL: https://microbenotes.com/
4. bioninja.com.au
This website is designed specifically for the new IB Biology syllabus (2016 –). Here you can find Interactive presentations for all SL and AHL topics, Downloadable slideshows (with optional narrations), Topic-specific activity sheets (with answers) and Summary notes for every core topic.
Website URL: https://ib.bioninja.com.au/
5. biologydiscussion.com
This website’s mission is to provide an online platform to help students to share notes in Biology. This website includes study notes, research papers, biology essays, articles, and other allied information submitted by visitors.
Website URL: https://www.biologydiscussion.com/
6. microbiologyinfo.com
MicrobiologyInfo.com is a constantly growing and evolving collection of microbiology notes and information. Whether you’re a student, professor, working in the medical field, or just curious about microbiology, you’ll find the articles interesting and informative. Writing comprehensive and accurate medical notes is a very time-consuming process, but this website tries to keep the site updated with the latest developments in the microbiology field.
Website URL: https://microbiologyinfo.com/
7. microbeonline.com
Microbeonline.com is an online guidebook of Microbiology, precisely speaking, Medical Microbiology. In this blog, we can find information and resources about pathogenic bacteria, viruses, fungi, and parasites. This website tries its best to make this site user-friendly and resourceful with timely/updated information about each pathogen, disease caused by them, pathogenesis, and laboratory diagnosis. You can also find Multiple Choice Questions (MCQs) in Bacteriology, Virology, Mycology, Parasitology, and Immunology sections.
Website URL: https://microbeonline.com/
8. biologycorner.com
The Biology Corner is a resource site for biology and science teachers and students. It contains a variety of lessons, quizzes, labs, web quests, and information on science topics for all levels, including introductory life science and advanced placement biology. You can find lessons related to biology topics in the links listed under “topics” on the sidebar as well as specific curriculum for biology classes taught in high school.
Website URL: https://www.biologycorner.com/
9. microbiologynote.com
Microbiologynote.com is an educational website for academic and research resources for students, researchers, and tutors in microbiology . Microbiologynote.com was founded in 2020 by Sourav, with the sake to provide information and reference material on Microbiology to students and professionals. Their mission is to make the best quality lecture notes in all areas of microbiology, and keep their website users up to date and informed with scintillating microbiology discoveries, news, resources, and other information for self and professional development.
Website URL: https://microbiologynote.com/
10. onlinebiologynotes.com
Online Biology Notes is the website founded by Gaurab Karki that provides biology notes on cell biology, microbiology, biochemistry, immunology, and biology practicals.
Website URL: https://www.onlinebiologynotes.com/
11. bioexplorer.net
Bioexplorer.net caters to the needs of students & teachers by providing complex biology topics in easy-to-understand formats with visuals, research articles, and also current biological news for free. This website shares breaking science news and articles on a variety of topics from the leading universities and research institutions around the globe. It is the ultimate guide to Biological Web Resources ranging from DNA to Plants & Animals and everything in-between. This site was originally developed for the researchers involved in biological studies but they want to make the science more enjoyable to even a common man too.
Website URL: https://www.bioexplorer.net/
12. thebiologynotes.com
The Biology Notes is an educational niche website related to biology (microbiology, biotechnology, biochemistry, zoology, botany, cell biology, genetics, molecular biology, etc.) and different other branches of biology. It was created on 2019-05-10 with the aim to provide biology notes for undergraduate and graduate students with the help of notes provided by the students and professionals in this field.
Website URL: https://thebiologynotes.com/
13. microscopemaster.com
MicroscopeMaster is a rapidly growing website with new articles posted every week! On this website. we can find articles on cell biology, the microbiology of our environment, experiments, all types of techniques and applications as well as up-to-date microscopy news to expand the knowledge. There are a wide variety of imaging techniques used in biological research as well as by the hobbyist or student, from the more basic to advanced, each with its pros and cons. MicroscopeMaster expands upon them – brightfield, darkfield, phase contrast, fluorescence, and more, to help you correctly determine which module is best to use in conjunction with your microscope. MicroscopeMaster loves to contribute to the education of children at every grade level. Importantly, a section focusing on student/kids models is quickly developing. There are several sturdy brands (Omax, Amscope, Celestron, Levenhuk, etc.) available that are perfect for your budding scientist as well as for your college student. Current information, reviews , and comparisons can help you find a good quality microscope in your price range, therefore depending on your needs, it may not be necessary to purchase the most advanced and so keep within your budget.
Website URL: https://www.microscopemaster.com/
14. alevelbiology.co.uk
This website provides A-Level Biology students and teachers with resources for use with the AQA, OCR, CIE, Eduqas Edexcel A/B & International exam boards. The resources can be used to aid teachers with their teaching in the classroom and students with their learning. Their A-Level Biology teachers have created all of the learning materials you need to pass your exams including Condensed revision notes aligned to AQA, OCR, CIE, Eduqas Edexcel A/B & International exam boards, Comprehensive exam question booklets for each exam board, 1200+ quickfire revision quizzes to practice knowledge, 54 mind maps to help you visualize the complex topics and Past papers directly from the exam boards.
Website URL: https://alevelbiology.co.uk/
15. easybiologyclass.com
Easy Biology Class provides an opportunity to master different fields of Biology and to prepare for different competitive examinations in the field of Life Sciences; thereby it enlightens about the enormous scope of biology and leads to different openings for higher studies and brighter careers. It is a resource site for biology lovers especially students and teachers who wish to have a good grip over biology in an easy and enjoyable way. On this website, you can find free biology tutorials, video lectures, lecture notes, PPT presentations, practical aids, mock tests, MCQs, Question Bank, and biology animations from different disciplines of biology.
Website URL: https://www.easybiologyclass.com/
16. biologyreader.com
Biology Reader is a one-stop destination for content seekers belonging to different fields of biology which includes science, microbiology, biotechnology, zoology, botany, and chemistry. Biology Reader has been started to deliver a concise summary of the mass information to the readers. This site mainly encourages the biology students who suffer during their exams, practicals, and presentations for well-organized study material. The motto of this website is ‘serving quality and comprehensive matter’. Biology Reader is a key solution for those who look forward to clarifying their doubts by facilitating in-depth knowledge, some interesting facts, comparative charts, diagrams, examples, and tabular presentations as a source of “Proper notes”. This makes their content more engaging, attractive, and reader-friendly.
Website URL: https://biologyreader.com/
17. notesonzoology.com
NotesOnZoology.com is an online platform to help students to discuss anything and everything about Zoology. This website includes study notes, research papers, essays, articles, and other allied information submitted by visitors.
Website URL: https://www.notesonzoology.com/
18. microbiologynotes.org
This website provides notes on different brances of microbiology.
Website URL: https://microbiologynotes.org/
19. biologyexams4u.com
This is a biology information site for everyone who loves and wishes to explore ‘the science of life’. Specifically, this site is meant for students pursuing higher studies in different streams of life science. Biology Exams 4 U provides details on the list of the exam you can apply in different streams of life sciences, the syllabus at a glance, the highlight of exams, simplified notes with key points, biology practice test and quizzes, MCQs, videos, and many more.
Website URL: https://www.biologyexams4u.com/
20. laboratoryinfo.com
LaboratoryInfo.com was launched on 23 March 2015 with the aim to provide every possible information, guide, and update related to Medical Laboratory Science to the students as well as laboratory professionals. This blog covers several disciplines primarily including Microbiology (Bacteriology, Virology, Mycology, Immunology, Parasitology), Biochemistry, Hematology, Histopathology, and Cytopathology.
Website URL: https://laboratoryinfo.com/
21. biodifferences.com
On this website, you can find biology differences on different terms related to Biochemistry, Botany, Ecology, Microbiology, General Science, Zoology, etc.
Website URL: https://biodifferences.com/
22. paramedicsworld.com
Paramedics World was founded in 2017 by Sahil Batra, with the sake to provide medical and paramedical information and reference material to the students and the professionals. The Blog is basically devoted to the Paramedical personnel who risk their lives to save the life of other people.
Website URL: https://paramedicsworld.com/
23. microbiologyinpictures.com
This website provides pictures of different bacteria on different culture media along with brief information.
Website URL: https://www.microbiologyinpictures.com/index.php
24. biologyjunction.com
Biology Junction is an essential site dedicated to helping those doing advanced placement biology work in high school. This site provides the best AP Biology review content and provides helpful information for both students and teachers of the subject. Biology Junction has a dedicated team of specialists whose mission in life is to turn AP Biology from what some would call a lackluster science lecture series into a more proactive learning experience. Through many years of time, they have developed and adopted the best of the best in terms of study aids, essential work guides, systems, and peripherals.
Website URL: https://www.biologyjunction.com/
25. learn-biology.com
This website is designed by a teacher, Glenn Wolkenfeld (Mr. W.), who can support you in achieving biology success for your own students. The quizzes, flashcards, and interactive diagrams in the tutorials on learn-biology.com mirror the techniques he uses in his classroom to guide his students toward success. He uses these interactive tutorials with his own students. Hundreds of teachers and professors are using them with thousands of students around the world.
Website URL: https://learn-biology.com/
26. microbiologynutsandbolts.co.uk
The purpose of this site, “Microbiology Nuts and Bolts” is to provide a clinically focused, no-nonsense guide to the key elements of microbiology and infection. These must-have resources are intended to stop common and often unnecessary mistakes that occur in everyday medicine and antibiotic prescribing. Written by a Consultant Clinical Microbiologist, they aim to provide doctors and healthcare staff with the ability to confidently identity the microorganisms that are the cause of a patient’s infection and how to treat them.
Website URL: http://www.microbiologynutsandbolts.co.uk/
27. onlinesciencenotes.com
Online Science Notes is the website founded by Sushil Humagain that provides science notes on physics, chemistry, biology, microbiology, biochemistry, immunology, geology, astronomy, etc.
Website URL: https://onlinesciencenotes.com/
28. biology4kids.com
It’s not just biology for kids, it’s for everyone. If you are looking for basic biology help and information, this is the site. They have information on cell structure, cell function, scientific studies, plants, vertebrates, invertebrates, and other life science topics. If you’re not sure what to click, try their site map that lists all of the topics on the site. If you get lost in all of the information, use the search function at the top or bottom of each page.
Website URL: http://www.biology4kids.com/
29. biologyeducare.com
Biologyeducare.com is a biology topic-related website that provides information for college and university students about Life Science which helps to learn about all sorts of biology terms, principles, and all types of life forms. Generally, they provide the scientific study of living organisms both plants and animals. They try to make the study easy and interesting for you. If you would like to gather basic biology knowledge, you should stay on this site “Biologyeducare.com” and you know and congregate any information about any biology-related topics for your needs.
Website URL: https://biologyeducare.com/
30. themedicalbiochemistrypage.org
The Medical Biochemistry Page is a portal for the understanding of biochemical, metabolic, and physiological processes with an emphasis on medical relevance. The Medical Biochemistry Page has been continuously updated and expanding, a free educational resource on the internet since 1996. The goal of the site is to provide extensive, detailed, and accurate information on a range of topics centered on the foundation of Medical Biochemistry. In addition to content that would be found in most Medical Biochemistry textbooks, The Medical Biochemistry Page contains integrated content related to physiology and pharmacology.
Website URL: https://themedicalbiochemistrypage.org/
31. rsscience.com
Their website’s mission is to provide good quality scientific kits without costing you lots of money and help the next young generation explore the beauty of the tiny world with clear instruction! A picture is more than a thousand words. You will find that they used many pictures, images, cartoon illustrations, and diagrams to explain scientific facts and principles.
Website URL: https://rsscience.com/
32. biotechnologynotes.com
This website provides biotechnology-related notes.
Website URL: https://www.biotechnologynotes.com/
33. bacteriainphotos.com
This website provides a photo gallery of medically important bacteria along with their characteristics.
Website URL: http://www.bacteriainphotos.com/index.html
34. biology-pages.info
This website represents an online biology textbook by John W. Kimball. Although some of the information has been drawn from the sixth edition of the author’s text Biology published in 1994 by Wm. C. Brown, every effort has been made to adapt the material to the opportunities provided by an online text. John W. Kimball has retired from a lifetime of teaching biology. A graduate of Harvard College, he began his teaching career at the secondary level, teaching chemistry and biology to students at Phillips Academy, an independent school in Andover, Massachusetts. In 1969, he returned to Harvard to study immunology with the late Professor A. M. Pappenheimer. After receiving his Ph.D. there, he went on to teach introductory biology (in both majors and nonmajors courses) and immunology at Tufts University where he became a tenured professor. In 1982 he returned again to Harvard where he taught immunology and also participated in teaching the introductory course for majors.
Website URL: https://www.biology-pages.info/
35. biomadam.com
This website provides notes on Biology and Health Science.
Website URL: https://www.biomadam.com/
36. biologyease.com
Biologyease is a biology niche educational website founded on May 20, 2020, to provide free quality notes, articles, and quizzes for various competitions. We provide notes for Microbiology, Biotechnology, Virology, Immunology, Zoology, etc.
Website URL: https://biologyease.com/
37. microbiologynotes.com
This website provides notes on Bacteriology, Virology, Parasitology, Mycology, and Laboratory Sciences.
Website URL: https://microbiologynotes.com/
38. overallscience.com
This website provides online Science Notes for Students.
Website URL: https://overallscience.com/
39. biosciencenotes.com
This website provides notes on biological sciences.
Website URL: http://www.biosciencenotes.com/
40. biologyreference.com
This website provides information on biology terms.
Website URL: http://www.biologyreference.com/
41. thebiologyprimer.com
The Biology Primer website provides courses and video lectures related to biology. Students are guided through the exercises reinforcing lecture concepts by incorporating interactive questions, diagrams, and charts.
Website URL: https://thebiologyprimer.com/
42. thesciencenotes.com
The Science Notes is a complete and reliable website that contains simple and understandable notes and references of all the faculties of Science. It aims to help students of all levels with a better understanding of Science.
Website URL: https://www.thesciencenotes.com/
43. microbe.net
The microBEnet website is a hub for information about the microbiology of the built environment. The microbiology of the Built Environment network (aka microBEnet) is a project funded by a grant from Alfred P. Sloan Foundation’s Program on the Indoor Environment to Jonathan A. Eisen at the University of California, Davis. The main goals of the microBEnet project are to Catalyze communication and collaboration among researchers funded in the Sloan Program on the Indoor Environment, Reach out to researchers in related fields (e.g., microbial ecology, building sciences) and provide them with resources that would help them begin to study the microbiology of the built environment and provide outreach to “stakeholders” outside of these fields (e.g., the general public, funding agency representatives, government staffers).
Website URL: https://microbe.net/
44. awkwardbotany.com
This website is founded by Daniel, and he is a bona fide plant nerd. He grows plants, study plants, work with plants, and write about plants. This blog will document his plant obsession. It is for the equal plant-obsessed, as well as for the plant interested and the plant curious. Along with being consumed with plants and the plant world, he is also immersed in awkwardness, bursting at the seams with geekiness, Hence, Awkward Botany.
Website URL: https://awkwardbotany.com/
45. wildlifebiology.org
Wildlife Biology is an Open Access journal. Wildlife Biology is a high-quality scientific forum directing concise and up-to-date information to scientists, administrators, wildlife managers, and conservationists. While Wildlife Biology primarily publishes ecological papers, contributions to the human dimensions of wildlife management are also welcome.
Website URL: https://www.wildlifebiology.org/
Below are the websites ranking from 46 to 61 with less than 5000 monthly visitors.
61. 24hoursofbiology.com
24HoursOfBiology is an electronic platform built to help the Biological community including students, teachers, and researchers out there. Their goal is to provide amazing high-quality contextual content with infographics and GIFs in a non-complicated manner that anybody can read and understand easily. Their content will possess all biology topics from basic to advance levels which will quickly register in the reader’s mind.
Website URL: https://24hoursofbiology.com/
61. askmicrobiology.com
Ask Microbiology not only contains Questions and Answers but also provides a platform to share knowledge with the world via blogging. Be a part of this community built exclusively for Microbiology.
Website URL: https://askmicrobiology.com/
61. biologybrain.com
Biologybrain.com site provides you with clear information about biological mechanisms which take place in living cells like prokaryotic cells and eukaryotic cells. One of their section (Biology topics) is mainly focusing on biology mechanisms that contain concept-oriented activities in different subjects. The subject-wise information provided on this site can help you to clear the competitive exams in life sciences like CSIR-JFR/NET, ICMR-JRF, DBT-JRF, ARS-NET, GATE, NEET, and state-wise entrance exams for lectureship or research. In their current science section, they regularly update various notifications like the novelty of research in national and international wise and information about scientific news and job opportunities.
Website URL: https://www.biologybrain.com/
61. biologyguide.app
BiologyGuide was founded by Simon Bluhm, BSc(Hons) MBChB FRCA in 2004, and has grown to be one of the best revision websites for AS and A level Biology students in the UK. BiologyGuide was recommended by the Guardian in December 2007.
Website URL: https://biologyguide.app/
61. biologyideas.com
This website helps you learn about all sorts of biology terms, principles, and life forms.
Website URL: https://biologyideas.com/
61. biologynotes.site
Biology notes are the one-stop platform that provides quality education. They want to promote free quality education throughout the world.
Website URL: https://www.biologynotes.site/
61. biopharmanotes.com
This website provides notes on biomedical and pharmaceutical sciences.
Website URL: https://biopharmanotes.com/
61. concisebiology.com
Concisebiology.com is a team of graduates in different fields of biology. Their goal is to provide concrete information to individuals through the articles. They also aim in helping everyone to gain the desired knowledge in the vast field of biology. They are constantly working towards improving and upgrading the contents and making the page as user friendly as possible.
Website URL: https://concisebiology.com/
61. generalmicroscience.com
This website provides notes on microbiology topics.
Website URL: https://www.generalmicroscience.com/
61. learnbiology.net
This website provides A-level biology revision lessons suitable for ALL Exams Boards. Many of the revision lessons are also suitable for BTEC Level 3 Nationals in Applied Science, International Baccalaureate (IB), Access to Science courses, and foundation degree – In Fact ANY Level 3 Biology! All the A-level biology revision lessons and resources have been made by an experienced biology lecturer with over 15 years of teaching experience. So, you can rest assured you’re getting high-quality resources that will help you learn, master, and succeed in your A-level biology revision and really help get you prepared for those dreaded exams.
Website URL: https://www.learnbiology.net/
61. microbes.info
Microbes.info is an internet website designed to bring useful and interesting microbiology informational resources to you. This website attempts to reduce the clutter and the size of the haystack in an effort to help you filter through the information in an organized manner. Microbes.info went live in mid-2002. The site is free to access. There are no costs for adding microbiological resources (microbiology sites, organizations, companies, journals, conferences, meetings, job postings , workshop postings, etc.).
Website URL: https://microbes.info/
61. microbesinfo.com
This website provides information on microbiology and infectious diseases founded by Prasil Pradhan.
Website URL: http://microbesinfo.com/
61. microbialfacts.com
Microbial Facts is a microbiology blog. It covers all the things about microbiology. It was launched on 5 November 2018. Microbial facts are the facts of viruses, bacteria, fungi, algae, and parasites. It discusses all the topics that include the activities of microorganisms, microbial diseases, antibiotic resistance, antibiotic alternatives, microbial cancer, and relevant topics.
Website URL: https://www.microbialfacts.com/
61. microbialmenagerie.com
The Microbial Menagerie is a website founded by Jennifer, a microbiologist, science communicator, and photographer. Jennifer created this blog in order to both educate in a variety of microbiology topics and bring attention to how microbiology affects our lives in ways we may not have thought about before.
Website URL: https://microbialmenagerie.com/
61. microbiologymatters.com
This is a website that is designed to be a resource site for people interested in microbiology and in particular those who have or are pursuing a career in any aspect of microbiology, including bacteriology, mycology, virology, and molecular biology.
Website URL: https://microbiologymatters.com/
61. sciencevivid.com
Sciencevivid is a specialized educational blog/website about several fields of biology (including microbiology, molecular biology, biochemistry, genetics, cell biology, and clinical genetics). Additionally, it provides the most recent and up-to-date news in technology, science, and research. It was founded on May 4, 2022, with the intention that undergraduate and graduate students (B.Sc, M.Sc, M.Phil, and Ph.D.) would have access to study notes and be able to comprehend them appropriately.
Website URL: https://sciencevivid.com/
Disclaimer: The information on these websites is taken from their respective website links. If there are any copyright issues regarding the information of the websites or if you want to edit the description of the website, or you want to list your website on this page, please email us at microbenotes(at)gmail(dot)com .
About Author
Sagar Aryal
1 thought on “Top and Best Biology Websites or Blogs of 2023 for Study Notes”
HI, I read this article which was very informative and helpful for student. Wonderful blog I recommended it to m students Thanks!
Leave a Comment Cancel reply
Save my name, email, and website in this browser for the next time I comment.
Biology Games & Virtual Labs!
Body systems (physiology), evolution & classification, genetics & meiosis, life chemistry (dna, proteins, etc,), respiration & photosynthesis, scientific methods, new ad-free membership site, ad-free member benefits.
- Improved Performance!
- Less Distraction!
- Build Your Own Games (coming soon)!
- Membership to BrainyBlast included (coming soon)!
Please note: There will always be a FREE version of BioMan Biology that has all of the same games, activities, and videos as the membership site, but I also wanted to offer an ad-free option for those who want to have it! To access the ad-free member site, click the "Member Site" button at the top left of this webpage!
Click the button below to see how you can become a member!
NEW! YouTube Channel
What is BioMan Biology?
BioMan Biology is the fun place to learn Biology! Here you will find learning games, review games, virtual labs and quizzes that will help you to learn about cells, ecology, genetics, physiology, and much more! Note: If you are a teacher, please check out the teacher section for ways to use the site to increase student engagement and learning. Remember, everything on this site is completely FREE to use!
Biology Video Games and Virtual Labs by Topic

Evolution and Classification
Genetics and meiosis.

Life Chemistry (Biochemistry and Molecular Biology)

Photosynthesis and Respiration
Scientific methods (investigation and experimentation ).

Privacy Policy
- Grades 6-12
- School Leaders
Get our FREE Classroom Seating Charts 🪑
Best Science Websites for Middle and High School
Biology, chemistry, physics, and beyond.

Science is exciting. Unfortunately, students can find the lessons a little dry. Whether you’re in the classroom or teaching online, finding the right resources can bring these complex concepts to life! To help you get started, here’s a list of the best science websites for middle and high school. Jump to your field of study:
- Earth Science
- Environmental Science
Best Science Websites for Teaching Biology
Hhmi biointeractive.
You may be familiar with HHMI’s free movies and posters; they also offer films that are available to stream from the site. Other options include 3-D interactives, virtual labs, and printable activities.
Biology Junction
If you need a template for lab reports, ideas for your biology club, pacing guides or lessons for biology, Pre-AP Biology, or AP Biology, this is a good place to start.
Biology Corner
Developed by a high school teacher, Biology Corner includes curated resources from around the web paired with extra practice and presentations and as well as ready-to-use investigations.
Virtual Urchin
Sounds odd, but this robust site hosted by Stanford University utilizes sea urchins as an engaging entry point to life science concepts ranging from basic biology (introductory microscopy and predator-prey relationships) to university-level curriculum (gene function in embryos).
This site’s evolution lab makes phylogeny and evolutionary history accessible to all students while scaffolding an understanding of the fossil record, the role of DNA in evolution, and an introduction to biogeography. Kids can also play the role of a molecular engineer by solving RNA folding puzzles.
National Geographic Education
The resource library offers learning materials and activities on topics such as Oceanography, Cloning, Heterotrophs, and Genetically Modified Organisms.
Annenberg Learner Interactives
Rediscovering Biology: Molecular to Global Perspectives is an advanced course designed for high school teachers who possess substantial knowledge of basic biology but who want to update their content knowledge and understanding. The multimedia course materials include video, online text, interactive web activities, and a course guide.
[contextly_auto_sidebar]
Best Science Websites for Teaching Chemistry
Ward’s science featuring ward’s world.
Check out Ward’s World, a new destination that offers middle schoolers and their teachers free classroom activities, how-to videos, tips, tricks, and resources that make science easier—and more fun! Find chemistry, biology, physics, and earth science.
ChemCollective
Like most chemistry sites, virtual labs and lesson plans are freely available, but ChemCollective stands out with their scenario-based activities and forensics tie-ins with activities like the “ Mixed Reception ” Murder Mystery.
Bozeman Science
Want clear, standards-aligned videos? If so, Bozeman Science is a great resource for teaching AP Chemistry. You’ll be able to flip your classroom and provide additional support to your students.
American Association of Chemistry Teachers
One of the best resources for Chemistry teachers nationwide, the AACT produces consistently high-quality resources, including labs, demonstrations, and activities. The best part is that their materials are organized by grade and topic.
Middle School Chemistry
Don’t let the name fool you. Sure, this site is perfect for middle school learners, but if you teach introductory chemistry or physical science, the level of materials is perfect for grades 9-10, as well. Lesson plans are easy to find, and some are even available in Spanish for English language learners!
The Periodic Table Interactive will take students through the periodic table piece by piece to give them a better understanding of how it works. Chemistry: Challenges and Solutions is a video instructional series on basic chemistry concepts and science history.
Chemdemos are virtual interactives for more advanced chemistry students. Particulate models and real-time data help you get through labs that you may not have sufficient materials to complete. They can also give your students extra practice at home before or after “wet” labs.
Molecular Workbench
This site facilitates a microscopic understanding of our macroscopic world. You’ll be blown away by resources like their Semiconductor and Chemical Bonding Module. All of the modules contain embedded assessments to keep your students on track and inform you of their progress.
ChemMatters Online
Always free for everyone, this is a terrific resource for middle school and high school science teachers as well as parents. Each issue provides a new collection of articles on chemistry topics that students will find engaging and relatable. The back-issue online library offers interesting downloadable articles on all sorts of chemistry-related topics, while the Teacher’s Guides help you direct your students as they learn from their reading.
Best Science Websites for Teaching Earth Science
The Dynamic Earth Interactive takes students through a visual feast through Earth’s layers and plate tectonics. The lessons can be extended by incorporating the Rock Cycle and Volcanoes Interactives.
National Oceanic and Atmospheric Administration Resource Collections
Along with oceans and coasts, weather, and more, instructors can find lesson plans in this collection that contain NOAA data and real-time weather information.
GeoInquiries
This collection includes all the major map-based concepts found in a typical middle or high school earth science course—topography, earthquakes, volcanoes, oceans, weather, and climate.
These digital labs focus on earth science topics for both middle and high school. Topics include continental plate boundaries, orbital patterns, and the sun-moon-earth system. All of their labs have automatic assessments to help teachers keep track of student growth.
High-Adventure Science
These free online curriculum lessons were developed for five days of classroom instruction and include one or more Earth systems models plus assessment items.
This resource library includes lessons covering a wide range of topics, including The Water Cycle, Erosion, Precipitation, and Metamorphic Rocks.
Best Science Websites for Teaching Environmental Science
Global footprint network ecological footprint calculator.
If you’re among those who calculate the ecological footprints as an activity in your AP Environmental Science class, you’ll enjoy this site. The questions are relatable to daily life, and the quiz can be retaken to analyze how lifestyle changes impact our footprint.
Population Education
This site helps to build a firm foundation so you can fully explore pollution, ecology, and biodiversity. It boasts interactive maps , a Find a Lesson feature, and learning materials available in Spanish.
National Energy Education Development Project
This great resource provides accurate energy usage data and information on energy technological advances through games, kits, math extensions, and their free downloadable Energy Infobooks .
The Habitable Planet: A Systems Approach to Environmental Science is a video course that explores the natural functions of Earth’s systems and Earth’s ability to sustain life. Earth Revealed is a video instructional series on geology for high school classrooms that shows the physical processes and human activities that shape our planet.
This collection supports the map-based concepts found in high school environmental science like speciation, pollution, population ecology, and energy.
Best Science Websites for Teaching Physics
phet interactive simulations.
Complete with teacher-submitted and reviewed lessons, these activities help students explore topics including circuits , waves , and quantum mechanics .
The Physics Classroom
With a curriculum corner, question bank, lab area, and a NGSS-devoted page, this is valuable resource is among the best science websites for K-12—including distance learning!
Share My Lesson
Search through this great collection of hundreds of categorized ready-to-use handouts, labs, and lectures submitted by physics teachers. You’ll find resources on the conservation of energy and momentum, electromagnetism, fluid mechanics, and more!
Flipping Physics
The content from this popular site is comedic, clear and includes helpful algebra and calculus reviews. That means you can focus on the science content without mathematical misunderstandings.
Physics for the 21st Century
This resource is a one-stop-shop for teaching—textbook included! You’ll find learning units, videos, interactive simulations, and even a comprehensive Facilitator’s Guide!
New Jersey Center for Teaching and Learning
A wealth of resources for physics, math, and chemistry online.
The Amusement Park Physics Interactive will help students explore how physics laws affect amusement park ride design. In this exhibit, they will have a chance to find out by designing their own roller coaster.
What science websites would you add to the list? Come share in our WeAreTeachers HELPLINE group on Facebook.
Plus, check out our lists of science books and steam apps ..

You Might Also Like
57 Tips, Tricks, and Ideas for Teaching 6th Grade
Brilliant ideas from brilliant teachers (like you). Continue Reading
Copyright © 2024. All rights reserved. 5335 Gate Parkway, Jacksonville, FL 32256
- International
- Education Jobs
- Schools directory
- Resources Education Jobs Schools directory News Search

** iGCSE Biology - Edexcel - OBSERVING PLANT & ANIMAL CELLS**
Subject: Biology
Age range: 14-16
Resource type: Lesson (complete)
Last updated
4 July 2024
- Share through email
- Share through twitter
- Share through linkedin
- Share through facebook
- Share through pinterest

Unlock Engaging Biology Lessons with Comprehensive Resources! Save precious planning time and elevate your classroom experience with our all-in-one Biology teaching package. This resource includes meticulously crafted lesson PowerPoints, engaging worksheets, and multiple practice question PowerPoints complete with model answers.
Designed to align with GCSE curriculum standards, these materials ensure comprehensive coverage of key topics. Ideal for busy teachers seeking high-quality, ready-to-use resources that facilitate effective learning and assessment.
Tes paid licence How can I reuse this?
Your rating is required to reflect your happiness.
It's good to leave some feedback.
Something went wrong, please try again later.
This resource hasn't been reviewed yet
To ensure quality for our reviews, only customers who have purchased this resource can review it
Report this resource to let us know if it violates our terms and conditions. Our customer service team will review your report and will be in touch.
Not quite what you were looking for? Search by keyword to find the right resource:
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- 10 July 2024
Serious errors plague DNA tool that's a workhorse of biology
- Katherine Bourzac
You can also search for this author in PubMed Google Scholar
Plasmids (shown here in a coloured transmission electron micrograph with various genes highlighted) are circular DNA structures used in biology laboratories. Credit: Dr Gopal Murti/Science Photo Library
Laboratory-made plasmids, a workhorse of modern biology, have problems. Researchers performed a systematic assessment of the circular DNA structures by analysing more than 2,500 plasmids produced in labs and sent to a company that provides services such as packaging the structures inside viruses so they can be used as gene therapies. The team found that nearly half of the plasmids had design flaws, including errors in sequences crucial to expressing a therapeutic gene. The researchers posted their findings to the preprint server bioRxiv last month ahead of peer review 1 .
The study shines a light on “a lack of knowledge” about how to do proper quality control on plasmids in the lab, says Hiroyuki Nakai, a geneticist at Oregon Health & Science University in Portland who was not involved in the work. He was already aware of problems with lab-made plasmids, but was surprised by the frequency of errors uncovered by the study. There are probably many scientific papers that have been published for which the results are not reproducible owing to errors in plasmid design, he adds.
Wasted time
Plasmids are popular tools in biology labs because bacteria, including the widely used model organism Escherichia coli , use the structures to store and exchange genes. This means that biologists can make designer plasmids containing various genes of interest, and then coax E. coli to take them up and make lots of copies.

Reproducibility crisis: Blame it on the antibodies
Bruce Lahn, chief scientist at VectorBuilder, a company based in Chicago, Illinois, that provides gene-delivery tools, says that he and other biologists have been noticing problems with plasmid quality for years. For example, when Lahn was a professor at the University of Chicago, a graduate student in his lab spent six months trying to reproduce two plasmids that had been reported in the scientific literature. “We didn’t think twice about the quality of the plasmids, but then the experiment wouldn’t work” because the plasmids contained errors, he says.
Now at VectorBuilder, Lahn says he’s faced with the issue all the time — so he decided to evaluate it systematically. When customers submit error-laden plasmids, “it ends up wasting a lot of time”, and the extra steps involved in doing quality control add to the cost of producing the plasmids and packaging them into viruses, he says.
The VectorBuilder team’s analysis found a hodgepodge of errors in the more than 2,500 plasmids it evaluated. Some contained genes that coded for proteins toxic to E. coli , which means that they could slow or stop the growth of the organisms biologists rely on to replicate their plasmids. Others, destined for packaging into viruses, encoded proteins toxic to those viruses. And some contained repetitive DNA sequences that can accumulate mutations inside plasmids.
Checking for errors
The most rampant errors Lahn and his colleagues found were related to a key gene-therapy tool. Therapies are often packaged into adeno-associated viruses (AAVs), which are mostly harmless and can ferry treatments to cells. When making the plasmids for these AAVs, researchers sandwich a therapeutic gene between sequences called ITRs, which play a crucial part in ensuring that the gene gets packaged into the virus for delivery. In essence, these sequences send a biological signal to cells that says “I belong in this virus”. But the team found that about 40% of the AAV plasmids in the study had mutations in the ITR regions that could garble this important message. If researchers were to use these misdesigned plasmids, their gene therapy might not work — and it could take the scientists a long time to find out why.
Engineered yeast breaks new record: a genome with over 50% synthetic DNA
Mark Kay, a paediatrics and genetics specialist at the Stanford School of Medicine in California, has also seen at first hand that plasmid errors can delay lab projects. But he’s confident that scientists can spot and fix these errors. He says that gene-therapy researchers are familiar with potential ITR issues, and that errors are unlikely to lead to problems in clinical settings. That’s because regulatory agencies such as the US Food and Drug Administration have stringent standards that require researchers to carefully analyse their plasmids before using them in the clinic.
Nakai says checking plasmids for errors by sequencing them could alert researchers to the problems highlighted in the study. A few companies, including Plasmidsaurus in Eugene, Oregon, and Elim Biopharmaceuticals in Hayward, California, offer plasmid sequencing for about US$15.00 per sample, says Nakai, who has no financial interest in either company. He also recommends that new lab members spend time learning from experienced plasmid constructors; it’s a tedious, artisanal process, he says, but if you get it wrong, it can waste a tremendous amount of time and money.
Another way for labs to avert issues is by publicly sharing their plasmid sequences in open-access repositories, says Melina Fan, chief scientific officer of the non-profit organization Addgene in Watertown, Massachusetts. Addgene provides one such repository, Fan says, and it “sequences the deposited plasmids and shares the sequence data via its website for community use”. Verification of plasmids is important, she adds.
Lahn hopes that his team’s analysis will draw researchers’ attention to the fact that these workhorse lab tools are often taken for granted. “The health of the tool is not something people question,” he says, even though they should.
doi: https://doi.org/10.1038/d41586-024-02280-1
Bai, X. et al. Preprint at bioRxiv https://doi.org/10.1101/2024.06.17.596931 (2024).
Download references
Reprints and permissions
Related Articles

- Gene therapy
- Scientific community
- Synthetic biology
- Biological techniques

Scientists edit the genes of gut bacteria in living mice
News 10 JUL 24
Streamlined collaboration can boost CRISPR gene therapies for rare diseases
Correspondence 02 JUL 24

Hope, despair and CRISPR — the race to save one woman’s life
News Feature 12 JUN 24

Edward C. Stone obituary: physicist who guided Voyager probes to interstellar space
Obituary 10 JUL 24

How PhD students and other academics are fighting the mental-health crisis in science
News Feature 09 JUL 24
Regulate to protect fragile Antarctic ecosystems from growing tourism
Correspondence 09 JUL 24

No CRISPR: oddball ‘jumping gene’ enzyme edits genomes without breaking DNA
Technology Feature 27 JUN 24
Selective haematological cancer eradication with preserved haematopoiesis
Article 22 MAY 24
Vaccine-enhancing plant extract could be mass produced in yeast
News & Views 08 MAY 24
Southeast University Future Technology Institute Recruitment Notice
Professor openings in mechanical engineering, control science and engineering, and integrating emerging interdisciplinary majors
Nanjing, Jiangsu (CN)
Southeast University
Neuroscience Research Assistant/Tech - Manhattan Weill Cornell Medical College
We are seeking motivated and enthusiastic research tech applicants to work on autism mouse models and brain organoids.
New York City, New York (US)
Weill Cornell Medical College
Postdoctoral Fellowship - Graph Database Developer
Postdoctoral Fellowship - Graph Database Developer Organization National Library of Medicine, National Institutes of Health, Bethesda, MD and surro...
Bethesda, Maryland
National Institutes of Health/National Library of Medicine
Postdoctoral Fellow - Boyi Gan lab
New postdoctoral positions are open in a cancer research laboratory located within The University of Texas MD Anderson Cancer Center. The lab curre...
Houston, Texas (US)
The University of Texas MD Anderson Cancer Center - Experimental Radiation Oncology
Senior Research Associates x 3 – Bioinformatician Team
The Genomics and Bioinformatics Core (GBC) within the Institute of Metabolic Science – Metabolic Research Laboratories at the Clinical School, Univers
Cambridge, Cambridgeshire
University of Cambridge
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies

Vision Neuroscientist Marie Burns Appointed Interim Director of the Center for Neuroscience

Interdisciplinary center is the heart of neuroscience research and training at UC Davis
- by Kimberly Cummings
- July 09, 2024
Marie E. Burns , a professor in the Departments of Ophthalmology and Vision Science, and Cell Biology and Human Anatomy in the School of Medicine, and a core faculty member at the Center for Neuroscience (CNS), has been appointed the CNS interim director, effective July 1, 2024.
Burns succeeds A. Kimberley McAllister , who has served as the center’s director since 2016 and who is departing UC Davis to begin a new position as Vice Provost for Research, Scholarly Inquiry and Creative Activity at Wake Forest University in the fall.
An accomplished vision scientist, leader and mentor
A highly respected and active member of the UC Davis neuroscience community, and a CNS core faculty member since 2001, Burns brings a wealth of leadership experience and a distinguished career in vision neuroscience research to her new role. She served as the inaugural director of the UC Davis Center for Vision Science from 2007–2012 and has been the director of the NIH Vision Science T32 Training Program since 2017.
“Marie’s dedication to mentoring trainees and fostering an inclusive, collaborative research environment has been a hallmark of her career at UC Davis,” said Mark Winey, dean of the College of Biological Sciences. “Her leadership acumen and longstanding commitment to advancing neuroscience research make Marie an excellent fit for this interim role.”

A third generation “eye doc,” Burns received her Ph.D. in neurobiology from Duke University and completed her postdoctoral training at Stanford University. “I am motivated by personal connections, my family members and friends affected by glaucoma and age-related macular degeneration,” said Burns. “I believe the work I do will directly impact the quality of care and treatment options within my parents' lifetimes."
Her research at CNS focuses on photoreceptors in the retina, the rods and cones in our eyes that detect light, in health and disease. The Burns Lab uses innovative approaches to study photoreceptor signaling and interactions between photoreceptors and the immune cells of the retina in vivo.
“Bridging a uniquely broad range of research topics and approaches, the Center for Neuroscience has been a national leader in multi-disciplinary neuroscience research for decades, and I’m honored to step into the role of interim director during this transition," said Burns. "Together with the incredible faculty, trainees and staff at CNS, we will continue to advance the center’s mission, building on the strong foundation established over the past 30 years.”
A decade of dramatic growth, innovation and discovery
McAllister, who has dual appointments in the Department of Neurology in the School of Medicine and the Department of Neurobiology, Physiology and Behavior in the College of Biological Sciences, is a cellular and molecular neuroscientist who specializes in synapse biology and neuroimmunology. Over nearly 25 years at UC Davis, McAllister has trained 10 pre-doctoral and 13 post-doctoral fellows and more than 60 undergraduates and 13 post-bacs. She has been a CNS core faculty member since 2000.
From 2014–2016, McAllister served as the center’s associate director and in 2016 became the first woman named director of the center. As director, McAllister oversaw significant growth in the center’s research funding, training programs, and philanthropic support, and played a pivotal role in fostering collaborations and interdisciplinary initiatives within the broad neuroscience community across campus.
“Kim’s tenure as CNS director has been marked by numerous significant achievements and contributions to our community, and to the broader community of neuroscientists at UC Davis,” said Winey. “Her legacy, which is one of achievement and innovation, of collaborative, interdisciplinary partnerships across our campus, will be enduring.”

During her tenure as director, McAllister expanded three research areas critical to the future of neuroscience discoveries: computational neuroscience, systems neuroscience and the field of neuroengineering. She also recruited seven CNS core faculty members, and greatly expanded NeuroFest , the center’s annual outreach event held each March during Brain Awareness Week. This event, which welcomes hundreds of visitors each year, has strengthened the center’s mission of engaging the public in neuroscience research. McAllister was also instrumental in creating the CNS Director’s Circle , a special recognition program for donors who support ground-breaking research, outreach events and training fellowships at the center.
McAllister is also the founding director of the UC Davis Learning, Memory, and Plasticity (LaMP) T32 Training Program, Co-Director of the UC Davis Conte Center and Co-Champion of the Emerging Health Threats Grand Challenge.
“Serving as the director of the Center for Neuroscience for the past eight years has been one of the greatest honors of my life,” said McAllister. “I am immensely grateful for my UC Davis colleagues and proud of all we have accomplished together.”
About the Center for Neuroscience
Established in 1992, CNS is the interdisciplinary hub for neuroscience research and training at UC Davis. CNS faculty are leaders in cell/molecular, computational, cognitive, development and systems neuroscience and conduct cutting-edge research across a wide range of neuroscience approaches and subfields. We are committed to training the next generation of neuroscientists and engaging the public in neuroscience research. Our commitment to diversity, equity, and inclusion permeates and elevates every aspect of our community.
Media Resources
- Kimberly Cummings, marketing and communications specialist, Center for Neuroscience, [email protected]
Primary Category
Secondary categories.

- Sign in / Register
- Administration
- Edit profile

The PhET website does not support your browser. We recommend using the latest version of Chrome, Firefox, Safari, or Edge.
- Search Menu
- Sign in through your institution
- Advance Articles
- Virtual Issues
- High-Impact Research Collection
- Celebrate 40 years of MBE
- Perspectives
Discoveries
- Cover Archive
Brief Communications
- Submission site
- Author guidelines
- Open access
- Self-archiving policy
- Reasons to submit
- About Molecular Biology and Evolution
- About the Society for Molecular Biology and Evolution
- Editorial Board
- Advertising and Corporate Services
- Journals Career Network
- Journals on Oxford Academic
- Books on Oxford Academic
Browse issues

Cover image

Volume 41, Issue 7, July 2024
Perspective, understanding the genetic basis of variation in meiotic recombination: past, present, and future.
- View article
Ancient and Recent Hybridization in the Oreochromis Cichlid Fishes
- Supplementary data
The Human Developing Cerebral Cortex Is Characterized by an Elevated De Novo Expression of Long Noncoding RNAs in Excitatory Neurons
Brain gene regulatory networks coordinate nest construction in birds, historic dog furs unravel the origin and artificial selection of modern nordic lapphund and elkhound dog breeds, morphometrics and phylogenomics of coca ( erythroxylum spp.) illuminate its reticulate evolution, with implications for taxonomy, evolutionary dynamics of accelerated antiviral resistance development in hypermutator herpesvirus.

Genomic Reconstruction of the Successful Establishment of a Feralized Bovine Population on the Subantarctic Island of Amsterdam
Selfing promotes spread and introgression of segregation distorters in hermaphroditic plants, deciphering structural traits for thermal and kinetic stability across protein family evolution through ancestral sequence reconstruction, genome-wide allele frequency changes reveal that dynamic metapopulations evolve differently, exploring the accuracy and limits of algorithms for localizing recombination breakpoints, multiple human population movements and cultural dispersal events shaped the landscape of chinese paternal heritage, rewinding the ratchet: rare recombination locally rescues neo-w degeneration and generates plateaus of sex-chromosome divergence, please mind the gap: indel-aware parsimony for fast and accurate ancestral sequence reconstruction and multiple sequence alignment including long indels, cmaple: efficient phylogenetic inference in the pandemic era, email alerts.
- Author Guidelines
Affiliations
- Online ISSN 1537-1719
- Copyright © 2024 Society for Molecular Biology and Evolution
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
share this!
June 27, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
Researchers propose a new, holistic way to teach synthetic biology
by Northwestern University
The field of synthetic biology, the science of manipulating biology, has a lot of "cooks in the kitchen," which has both helped it flourish and made it unusually difficult to create a cohesive, consistent curriculum for students at every level of study. Each discipline involved—from chemical engineering to ethics—has a unique approach to teaching and literature, which creates inconsistencies between what scientists learn.
Now, Northwestern University researchers propose a new way to teach synthetic biology that uses different levels of organization—starting at the molecular scale and growing to the societal scale—to teach core principles and a holistic view of developing sustainable synthetic biology technologies. The approach incorporates components from many disciplines, allowing people from many different backgrounds to access synthetic biology education.
A paper detailing the framework is published in the journal Nature Communications .
"Early versions of synthetic biology courses lacked a conceptual foundation," said Julius Lucks, a synthetic biology expert and lead author. "From an educational perspective, you saw a kind of hodgepodge depending on which department you were in. We set ourselves the challenge of trying to figure out: How can you merge all those disciplines, somehow develop a common framework and create a common language?"
Lucks is a professor of chemical and biological engineering at Northwestern's McCormick School of Engineering and co-director of the Center for Synthetic Biology (CSB).
When genetic engineering emerged in the 1970s, the idea to reuse, repurpose and reconfigure biological systems to address challenges in society—the core of synthetic biology—became possible. With CRISPR's advent around 1990, synthetic biology was popularized, and without any curriculum in place, faced an identity problem.
"One of the biggest problems we saw in our students and labs are that they tend to be focused on a very specific problem," said Ashty Karim, a Northwestern research assistant professor of chemical and biological engineering and the paper's first author.
"For example, if you're looking at how CRISPR works, you might be studying the localized protein machinery that makes edits to the DNA. But if you're going to create a technology based on CRISPR, there are many other important facets than the molecular scale workings. How does it work in the context of a cell or a population of cells within someone's body across different tissues? How does that interact with current health care systems? These are discussions that we need to be having and might influence the science that we do."
Karim is the director of research at CSB and a core member of its faculty.
From this came the idea of breaking down biology into scales, developed originally from a conversation about how most introductory biology courses present a continuum that moves from DNA to tissue to organism. There are emerging behaviors that appear at different levels of organization; society behaves differently from an organism, and so on. Synthetic biology, according to the paper, can be broken down into five components: molecular, circuit/network, cellular, biological communities and societal.
Key to the proposed framework is the presence of robust ethics at each scale.
By piloting the curriculum with undergraduate and graduate students at Northwestern, the authors said they found that different core principles, like thermodynamics and kinetics, mapped onto different scales naturally. For instance, do you need to understand a principle all the time? Or can you "ignore" it up to a certain scale?
Curriculum should also be underpinned by case studies that help learners analyze how engineering choices made at one scale affect biological function at another, assemble potential solutions to global challenges across scales and identify the impact of synthetic biology on societal goals and ethical issues. Case studies can also be tailored to the institution or instructor implementing the curriculum.
Presenting the approach to classes has been greatly successful, according to Lucks and Karim, who both said the concept "clicked" for students even the first time they taught the course. The scales concept has been so successful that in fact, the Northwestern Center for Synthetic Biology uses it to organize its cutting-edge collaborative research.
The researchers also hope the curriculum can be implemented much more broadly and have provided resources and ideas that others may use to adapt the approach to their needs and interests.
Journal information: Nature Communications
Provided by Northwestern University
Explore further
Feedback to editors

A new species of extinct crocodile relative rewrites life on the Triassic coastline
8 hours ago

New method achieves tenfold increase in quantum coherence time via destructive interference of correlated noise
9 hours ago

Mars likely had cold and icy past, new study finds

Study: Nanoparticle vaccines enhance cross-protection against influenza viruses

New tools are needed to make water affordable, says study
10 hours ago

Researchers demonstrate how to build 'time-traveling' quantum sensors

Lion with nine lives breaks record with longest swim in predator-infested waters
11 hours ago

New multimode coupler design advances scalable quantum computing

High-speed electron camera uncovers new 'light-twisting' behavior in ultrathin material

Perceived warmth, competence predict callback decisions in meta-analysis of hiring experiments
12 hours ago
Relevant PhysicsForums posts
Is meat broth really nutritious.
13 hours ago
Havana Syndrome
15 hours ago
Innovative ideas and technologies to help folks with disabilities
Jul 7, 2024
COVID Virus Lives Longer with Higher CO2 In the Air
Conflicting interpretations of rosemary oil study.
Jul 3, 2024
Who chooses official designations for individual dolphins, such as FB15, F153, F286?
Jun 26, 2024
More from Biology and Medical
Related Stories

Researchers create new tools to monitor water quality, measure water insecurity
Feb 17, 2020

Biosensors change the way water contamination is detected
Feb 8, 2023

Researchers develop 'founding document' on synthetic cell development
May 3, 2024
Researchers detect fluoride in water with new simple color change test
Jan 4, 2023

Chemistry as a building block for scientific literacy?
Jul 5, 2022

Simple test could prevent fluoride-related disease
Dec 16, 2019
Recommended for you

Subsurface of fingernails found to have precise tactile localization
16 hours ago

Two new species of Psilocybe mushrooms discovered in southern Africa
Jul 2, 2024

Unlocking biodiversity insights from the tropical Andes
Jun 27, 2024

Biomechanics of sound production in high-pitched classical singing
Jun 18, 2024

'Sour Patch' adults: 1 in 8 grown-ups love extreme tartness, study shows
Apr 29, 2024

Linking environmental influences, genetic research to address concerns of genetic determinism of human behavior
Feb 27, 2024
Let us know if there is a problem with our content
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Phys.org in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
Marcus Feldman receives lifetime achievement award from the Society of Molecular Biology and Evolution

Marcus Feldman , the Burnet C. and Mildred Finley Wohlford Professor and professor in the Department of Biology, has been selected as the 2024 winner of the Society of Molecular Biology and Evolution’s Lifetime Achievement Award. This honor recognizes Feldman’s outstanding research during his career and contributions to the field of molecular biology and evolution.
Read more about the award.
Photo courtesy of Stanford News Service
- Natural Sciences
© Stanford University, Stanford, California 94305
- Open access
- Published: 01 July 2024
Tagging large CNV blocks in wheat boosts digitalization of germplasm resources by ultra-low-coverage sequencing
- Jianxia Niu 1 , 2 ,
- Wenxi Wang 1 ,
- Zihao Wang 1 , 2 ,
- Zhe Chen 1 ,
- Xiaoyu Zhang 1 ,
- Zhen Qin 1 ,
- Lingfeng Miao 1 ,
- Zhengzhao Yang 1 ,
- Chaojie Xie 1 ,
- Mingming Xin 1 ,
- Huiru Peng 1 ,
- Yingyin Yao 1 ,
- Jie Liu 1 ,
- Zhongfu Ni 1 ,
- Qixin Sun 1 &
- Weilong Guo ORCID: orcid.org/0000-0001-5199-1359 1
Genome Biology volume 25 , Article number: 171 ( 2024 ) Cite this article
500 Accesses
Metrics details
The massive structural variations and frequent introgression highly contribute to the genetic diversity of wheat, while the huge and complex genome of polyploid wheat hinders efficient genotyping of abundant varieties towards accurate identification, management, and exploitation of germplasm resources.
We develop a novel workflow that identifies 1240 high-quality large copy number variation blocks (CNVb) in wheat at the pan-genome level, demonstrating that CNVb can serve as an ideal DNA fingerprinting marker for discriminating massive varieties, with the accuracy validated by PCR assay. We then construct a digitalized genotyping CNVb map across 1599 global wheat accessions. Key CNVb markers are linked with trait-associated introgressions, such as the 1RS·1BL translocation and 2N v S translocation, and the beneficial alleles, such as the end-use quality allele Glu-D1d (Dx5 + Dy10) and the semi-dwarf r-e-z allele. Furthermore, we demonstrate that these tagged CNVb markers promote a stable and cost-effective strategy for evaluating wheat germplasm resources with ultra-low-coverage sequencing data, competing with SNP array for applications such as evaluating new varieties, efficient management of collections in gene banks, and describing wheat germplasm resources in a digitalized manner. We also develop a user-friendly interactive platform, WheatCNVb ( http://wheat.cau.edu.cn/WheatCNVb/ ), for exploring the CNVb profiles over ever-increasing wheat accessions, and also propose a QR-code-like representation of individual digital CNVb fingerprint. This platform also allows uploading new CNVb profiles for comparison with stored varieties.
Conclusions
The CNVb-based approach provides a low-cost and high-throughput genotyping strategy for enabling digitalized wheat germplasm management and modern breeding with precise and practical decision-making.
Wheat is one of the most widely grown and consumed crops and provides 20% of the total protein and calories in human nutrition [ 1 ]. Accurate identification and evaluation of genomic polymorphism within wheat germplasm resources are crucial to enhancing breeding capacity and developing improved varieties with higher yields and resistance to biotic and abiotic stresses [ 2 , 3 ]. Beyond single nucleotide polymorphisms (SNPs) and small InDels, there are extensive structural variations (SVs) at a large scale in the wheat genome, which includes gene presence/absence variations (PAVs), copy number variations (CNVs), and chromosomal translocations, serving as an important source of genetic diversity in the wheat breeding population [ 4 , 5 , 6 ]. Current methods for identifying SVs generally require high-quality genome assemblies, high sequencing depth, or long-read sequencing [ 7 ], while the high cost of sequencing hinders profiling SVs at a population level.
DNA-based markers have been widely used for describing varieties and assisting breeding [ 8 ]. Multiple types of molecular markers derived from genomic variations have been developed to assist genome-based breeding in wheat, such as simple sequence repeats (SSRs), amplified fragment length polymorphisms (AFLPs), and SNPs [ 9 ]. Hybridization-based and PCR-based markers were the earliest molecular markers, which are time-consuming and laborious in genotyping, thus were difficult to apply in large-scale population analysis [ 10 ]. High-throughput methods such as genotyping-by-sequencing (GBS)-based and array-based SNP genotyping techniques were utilized to identify SNP/InDel markers [ 11 , 12 ], while these effective markers are still limited and the overall cost to genotype one sample is still high to be utilized in assisting the variety management and breeding design [ 13 ]. More stable and effective DNA markers and corresponding cost-efficient scanning methods are urgently needed for describing and exploiting wheat germplasm resources.
Common wheat is a typical allohexaploid crop, its genome is of considerable tolerance to large segmental deletions and duplications [ 14 , 15 , 16 ], and highly plastic to take in both intraspecific and interspecific introgressions [ 17 ]. Furthermore, the genomes of modern wheat germplasms have been shaped by introgression from wild relatives during domestication [ 18 ] and by distant hybridization during the modern breeding process [ 19 ]. Thus, the characterization of SVs in wheat is important for accurate genotyping of massive varieties. Reported cytogenetic and molecular methods for detecting SVs, such as fluorescence in situ hybridization (FISH) and genomic in situ hybridization (GISH), are limited in aspects of throughput and resolution and are primarily used to confirm known SVs [ 20 , 21 ]. Thus, there is an urgent need for a high-throughput and cost-effective method to characterize and exploit SVs across diverse wheat varieties.
The advent of the pan-genomic era brings opportunities for detecting SVs among wheat varieties globally [ 15 ]. Current main strategies include directly comparing genome assemblies and inferring SVs from the mapping of high-coverage resequencing data against the reference genome [ 22 , 23 ]. However, the high costs of sequencing-based strategies hinder the application in genotyping the SVs at a large-scale population level [ 7 ]. Recently, Keilwagen et al. demonstrated that the depth-of-coverage of GBS or low-coverage whole genome sequencing could be used for detecting large chromosomal variations [ 24 ]. However, there is still a lack of an accurate and cost-efficient method for characterizing depth-of-coverage variations with stable performance across wheat varieties, tackling the highly noisy signals introduced by low-coverage sequencing data.
Here, we identified a set of high-quality CNV block (CNVb) markers by tagging large CNV blocks from a worldwide collection of wheat resequencing data using a pan-genome reference, supporting accurate profiling of the CNVb markers across wheat varieties even at an ultra-low sequencing coverage. The link between in silico CNVb markers with key introgressions and benefit alleles associated with agronomic traits further adds value to the digitalization of wheat germplasm. A free-to-access and user-friendly web platform ( http://wheat.cau.edu.cn/WheatCNVb/ ) was also developed to help access and utilize the CNVb markers. In summary, the in silico CNVb markers can serve as new-generation molecular markers facilitating the characterization of the germplasm resources and assisting the genomic breeding in crops with high accuracy and low cost.
Pervasive large CNV blocks identified in wheat
To comprehensively survey and characterize the genome-wide copy number variations (CNVs) in wheat, we collected a panel of whole genome resequencing data of worldwide wheat accessions [ 16 , 17 , 25 , 26 , 27 ], including 186 modern cultivars and 342 landraces (Additional file 1 : Table S1). After mapping reads against the Chinese Spring (CS) reference genome IWGSCv1 [ 28 ], relative read depths were calculated bin-wisely with a bin size of 100 Kb. A total of 8430 Mb and 3375 Mb non-redundant bins were identified as deletion and duplication in at least one accession, respectively. The CNV bins were unevenly distributed across the genome, with higher frequencies observed near the ends of chromosomes (Additional file 2 : Fig. S1), consistent with observations in maize [ 29 ] and rice [ 30 ]. Our results showed that the total length of CNV regions ranged from 139 to 1567 Mb across different varieties (Fig. 1 a). Notably, the total lengths of CNV regions for 81.6% of the accessions exceed 500 Mb, with an average of 2061 CNV regions per accession, confirming that large CNV regions are pervasive across diverse wheat varieties. In contrast, maize and rice show fewer CNV regions, with average total lengths of 382 Kb and 142 Kb, and average counts of 53 and 19 CNV regions, respectively (Fig. 1 b), highlighting the high-frequent and large CNV blocks as a unique feature of wheat.

Characterization of large CNV blocks in wheat. a Distribution of total length and total count of CNV regions in each wheat accession. b Comparison of length and count of CNV regions between wheat, maize, and rice across the whole genome. c CNV region distribution of Jagger along chromosomes (1D, 2A, 2D, 5A, 5B, and 6B). Bin size, 100 Kb. d CNV region distribution on chromosome 1B among represent accessions (Lunxuan987, Aikang58, Mace, CDC Stanley, ArinaLrFor, and Julius). Bin size, 100 Kb. e Schematic representation of the conversion from the large CNV blocks (CNVb) (left) to tagged CNVb markers (right) on chromosome 6B. Left panel, CNV blocks with length ≥100 Kb. Right panel, CNVb markers, each color represents one unique CNVb marker. A particular CNV block (chr6B:265–278 Mb) was marked by a dashed rectangle. Bin size, 100 Kb
As a key feature of CNV regions detected in wheat compared with rice or maize, there are more CNV regions exceeding one megabase. There are six extra-large CNV blocks with lengths spanning ≥10 Mb detected on chromosomes 1D, 2A, 2D, 5A, 5B, and 6B in the wheat cultivar Jagger (Fig. 1 c). Many CNV blocks are shared among accessions, while positions and lengths of CNV blocks differ (Fig. 1 d, e). Taking chromosome 1B as an example, a large CNVb-deletion (chr1B: 0–236.7 Mb) across 1BS can be observed in both Lunxuan987 and Aikang58 (Fig. 1 d), which corresponds to the documented 1RS·1BL translocation [ 20 ]. Rather than the 1RS·1BL-related large CNV block presented in Lunxuan987 and Aikang58, several smaller CNV blocks were identified in Mace, CDC Stanley, ArinaLrFor, and Julius along the similar chromosome region (Fig. 1 d), reflecting the diversity of CNV blocks on the genome. The consistent local CNV block landscapes observed among accessions may serve as ideal markers to identify genetic relationships of wheat resources. Thus, we proposed a strategy to tag intraspecific shared CNV blocks as in silico markers for wheat germplasm identification. As a prototype example, we selected the large CNV regions spanning chromosome 6B shared among CDC Landmark, Jagger, ArinaLrFor, and Aikang58, and grouped them into five CNVb markers, which could effectively compress the comprehensive CNV landscapes into a list of digitalized signals (Fig. 1 e). The presence or absence of CNVb makers demonstrates an alternative and effective strategy in constructing the molecular fingerprints of wheat germplasm.
Developing CNVb markers at pan-genome level
To mitigate the potential bias of CNVb identification introduced by using a single reference genome and acquire sufficient markers, we constructed a pan-genome reference by iterative mapping of the whole-genome resequencing data against 16 de novo assembled reference genome sequences [ 15 , 26 , 31 , 32 , 33 , 34 , 35 ] (Additional file 1 : Table S2). This iterative process involved using a 1 Mb sliding window to identify sequences present in genomes other than Chinese Spring (CS). Starting with the Aikang58 genome as the initial reference, and progressing through each genome in order of assembly quality, we systematically detected and compiled 975 novel genome blocks with a total length of 2.7 Gb (Additional file 1 : Table S3). These blocks, which represent genomic regions absent in CS, were then assembled into a non-Chinese Spring pan-genome chromosome, denoted as “chrNCP” (Fig. 2 a). The saturation analysis showed that the number of non-CS sequence blocks increased by adding new assemblies and approached a plateau when 14 genomes were included (Additional file 2 : Fig. S2), indicating the representativeness of the constructed pan-genome by integrating a total of 17 wheat assemblies.

Development and evaluation of CNVb markers. a Pipeline to identify CNVb markers against wheat pan-genome. Pre-step, the pan-genome was constructed by combining the Chinese Spring assembly and the unmapped blocks of Chinese Spring relative to the other 16 wheat assemblies. Step 1, the initial CNV blocks of 528 high-quality wheat resequencing accessions were identified with a 100 Kb window. Step 2, a hidden Markov model (HMM) was introduced to smooth noisy signals, and then low frequency and short CNV blocks were filtered from the retained CNV blocks. CNV blocks with reciprocally overlapped regions larger than 80% were merged as a single CNVb cluster, and linkage clusters with close distances were further combined. Step 3, the final CNVb markers were extracted by eliminating those with low recall rates identified by ultra-low-coverage whole genome sequencing (ulcWGS), using CNVb markers identified by high-coverage whole genome sequencing as the ground truth. b Saturation analysis of CNVb markers. Five accessions were randomly added each time. The shaded area represents 100 replications for each sampling. The blue dot represents the average number of CNVb markers across 100 repetitions. c Comparison of the accuracy of SNPs, raw CNVs, and CNVb markers identified at low sequencing depth
To identify representative and stable CNVb blocks for distinguishing wheat accessions, we developed a pipeline to obtain high-quality CNVb markers by examining consistent borders, overlaping ratio, and continuity of CNV blocks based on a panel of whole genome resequencing data with an average coverage of 5.4×, which covers 528 wheat accessions (Additional file 1 : Table S1). The pipeline consists of three main steps: detecting raw CNV blocks, deducing low-confident and redundant CNV blocks, and removing CNV blocks sensitive to low sequencing depth (Fig. 2 a). Step 1, we mapped the sequencing data to the pan-genome reference and identified an initial set of CNV bins based on the read depth with a window size of 100 Kb. Step 2, we applied a hidden Markov model to generate continuous CNV blocks (Additional file 2 : Fig. S3), followed by the removal of short-length and low-frequency CNV blocks. CNV blocks sharing one border and have more than 80% overlapping regions were merged. CNVs closely linked in one cluster were further grouped. Then, an initial set of 8134 CNVb makers were identified genome-widely across the population. Step 3, we eliminated CNVb blocks with low recalls in ultra-low-coverage whole genome sequencing (ulcWGS) data (0.1×) to develop stable in silico markers (Additional file 2 : Fig. S4). Finally, our pipeline yielded a total of 1240 non-redundant high-quality CNVb markers, comprising 1045 CNVb-deletion markers and 195 CNVb-duplication markers. By profiling these CNVb markers across the genome, we observed that these CNVb markers are distributed across all chromosomes, with an average of 59 markers per chromosome (Additional file 2 : Fig. S5). We also showed that these CNVb markers span most regions of chromosomes, occupying up to 92.6% of each chromosome (Additional file 2 : Fig. S6), indicating that the developed in silico DNA marker set offers the feasibility of representing wheat genomic variation genome-widely. We further performed saturation analysis of CNVb markers and showed that 95% of CNVb markers could be recalled when the panel size reached 230 (Fig. 2 b), indicating the selected CNVb markers are sufficient for capturing the large CNV blocks at a population level in wheat.
High recalls achieved by scanning CNVb markers in ultra-low-coverage sequencing data
To evaluate the performance of scanning the in silico CNVb markers in new varieties, we examined the recalls of CNVb markers by scanning sequencing datasets at various coverages, which were randomly sampled from high-coverage resequencing data. The results showed that the performance of developed CNVb markers exceeded raw CNV regions and SNPs, especially for the ultra-low coverage data. The CNVb markers achieved a mean recall of 99.3% even at coverage of 0.05× (Fig. 2 c), highlighting the superiority of CNVb as a stable marker compared to traditional strategies. This result also suggests that CNVb could serve as a cost-efficient genotyping strategy for constructing DNA-based digitalized fingerprints of massive varieties based on ultra-low sequencing data.
Linking CNVb markers with known structural variations and beneficial alleles
To fully harness the potential of wheat germplasm carrying beneficial alleles such as ones conferring disease resistance for breeding applications, we linked numerous well-known structural variations and beneficial alleles with CNVb markers (Table 1 ). The r-e-z haploblock on chromosome 4B with approximately 500 Kb deletion, characterized by simultaneous absence of the Rht-B1 , EamA-B , and ZnF-B genes contributing to both the compactness and enhanced yield of semi-dwarf wheat [ 36 ], was digitized to a CNVb-deletion marker (CNVb.647, chr4B: 30.5–31.1 Mb) and identified in a total of 10 accessions (Fig. 3 a, Additional file 1 : Table S4). The pericentric inversion in chromosome 6B (perInv-6B), one of the most predominant chromosomal variants in wheat modern cultivars [ 21 ], was associated with a CNVb-duplication marker (CNVb.989, chr6B:167.9–183.4 Mb) on chromosome 6B. This association is based on the identification of a duplication block marker that is unique to varieties carrying the 6B inversion (Fig. 3 b). Our study identified 12 previously reported accessions harboring perInv-6B [ 20 , 21 ] and 21 additional accessions with the perInv-6B associated CNVb markers (Additional file 1 : Table S5). The high-molecular-weight glutenin Glu-D1d (Dx5 + Dy10) allele is associated with superior bread-making quality [ 37 ]. We developed a CNVb-deletion marker (CNVb.162, chr1D: 412.1–412.5 Mb) corresponding to the Glu-D1d allele (Fig. 3 c) and identified 11 accessions, such as Jagger, carrying the Glu-D1d allele (Additional file 1 : Table S6), which was proven by SDS-PAGE [ 37 , 38 ]. We identified 419 additional accessions that may carry the Glu-D1d allele (Additional file 1 : Table S6). These intriguing results indicate the current panel of CNVb markers could serve as an alternative way for efficiently scanning the presence of beneficial alleles or key structure variations among wheat varieties.

Associating CNVb markers with known structural variations and predominant haplotypes. a The CNVb-deletion marker CNVb.647 (chr4B: 30.5–31.1 Mb) corresponds to the r-e-z haplotype on chromosome 4B. b The CNV-duplication marker CNV.989 (chr6B: 167.9–183.4 Mb) is associated with the pericentric inversion on chromosome 6B (perInv-6B). c The CNVb-deletion marker CNV.162 (chr1D: 412.1–412.5 Mb) corresponds to the Glu-D1d (Dx5 + Dy10) allele of the high-molecular-weight glutenin gene Glu-D1 . d The CNVb-deletion marker (CNV.67.1, chr1B: 0–239.3 Mb) associates with the 1RS·1BL translocation. The “s1” denoted the subtype 1RS·1BL translocation associated with CNVb.67.1 (chr1B: 0–239.3 Mb). The “s2” denoted the subtype 1RS∙7DL/7DS∙1BL translocation associated with CNVb.67.2 (chr1B: 0–236.7 Mb). e The CNVb-deletion marker CNVb.290 (chr2B: 89.5–769 Mb) corresponds to the introgression from Triticum timopheevii on chromosome 2B. f Distribution of three types of CNVb allelic genotypes within the first 40 Mb region of chromosome 2A. The first type of CNV allele (CNV. 189, chr2A: 0–24.7 Mb) corresponds to the introgressed segment from Aegilops ventricosa . g – i PCR validation on three types of CNVb alleles located in the 25 Mb telomeric region of chromosome 2A. M, DNA marker 5000. Two primers were designed using partial sequences of the introgression fragments in Jagger ( g ) and Zang1817 ( h ), respectively, and the other was designed using partial sequences from 0 to 24.7 Mb on chromosome 2A of the CS genome ( i )
CNVb markers associated with reported interspecific introgressions in wheat were also annotated (Table 1 , Additional file 1 : Table S7). For instance, the well-documented 1RS introgression from rye ( Secale cereale ) carries multiple genes ( Pm8/Sr31/Lr26/Yr9 ) contributing to disease resistance in wheat. In this study, we developed a CNVb-deletion marker (CNVb.67.1, chr1B: 0–239.3 Mb) associated with 1RS based on the high sequence divergence between the 1RS and 1BS (Fig. 3 d). The CNVb.67.1 was identified in 16 accessions that were convinced as harboring the 1RS·1BL translocation by fluorescence in situ hybridization [ 20 , 21 ], including Lunxuan987. Additionally, we reveal there are 95 more accessions that also have the 1RS·1BL translocation-associated CNVb markers (Additional file 1 : Table S7). Our previous study showed the read depth pattern of 1B chromosome could well distinguish the two subtypes of 1RS-related translocation [ 16 ]. Based on the previous effort, we further developed a sub-CNVb marker on chromosome 1B (CNVb.67.2, chr1B: 0–236.7 Mb) in Aimengniu for representing the unique translocation (RT 1RS∙7DL/7DS∙1BL), which is different with the typical 1RS·1BL translocation in boundaries (Fig. 3 d and Additional file 2 : Fig. S7). Another example is the cultivar LongReach Lancer carrying two introgressed regions from different species, including a pericentric region spanning 427 Mb on chromosome 2B from Triticum timopheevii and a terminal segment of ~60 Mb on chromosome 3D from Thinopyrum ponticum [ 15 ]. Accordingly, the Triticum timopheevii related introgression is associated with CNVb markers CNVb.290 (chr2B: 89.5–769.0 Mb) (Fig. 3 e), and the Thinopyrum ponticum related introgression is associated with CNVb markers CNVb.540 (chr3D: 592.2–616.0 Mb, Additional file 2 : Fig. S8), respectively. By scanning our collection of wheat varieties, we showed the variety BAXTER also carries the Triticum timopheevii introgression (Fig. 3 e). Collectively, we exemplified that structural variations and introgression haplotypes could be transformed into digitalized CNVb markers with application potential for scanning larger wheat variety panels.
As pervasive independent introgressions have been utilized in modern wheat breeding, we further showed CNVb marker could distinguish introgressions even with overlapped genomic coordinates. We identified two CNVb markers at the end of chromosome 2A short arm. The CNVb-deletion marker CNVb.189 (chr2A: 0–24.7 Mb) was detected in Jagger (Fig. 3 f), which is linked to a 2N v S introgression from Aegilops ventricosa that conferred resistance to wheat blast and carried the rust disease resistance gene cluster ( Lr37/Yr17/Sr38 ) [ 15 ]. An additional 141 varieties were detected with the CNVb.189 marker (Additional file 1 : Table S8), such as Lankao198 (Fig. 3 f). The second CNVb-deletion marker overlapped with CNVb.189 is CNVb.173 (chr2A: 11.5–21.0 Mb), which was detected in Tibetan semi-wild wheat accession Zang1817 and Chinese cultivar Bima4 (Fig. 3 f). Collinearity analysis between Zang1817 and CS genome showed a degree of collinearity in the deletion region, despite low-quality alignment (Additional file 2 : Fig. S9), indicating that the CNVb.173 marker corresponds to an interspecific introgression. 50.9% of wheat varieties showed no CNVb maker detected in the first 25 Mbp regions of chromosome 2A, indicating three types of alleles as distinguished by CNVb markers. To validate the three identified alleles, we performed a PCR analysis by designing primers specific to 2N v S introgression sequences in the Jagger assembly (Fig. 3 g), to the sequences in the Zang1817 assembly (Fig. 3 h), and to the wild-type Chinese Spring assembly (Fig. 3 i), and results showed the yielded amplification in Jagger, Lankao198, Zang1817, Bima4, CS, and Aikang58 matched with the predicted allele types by CNVb markers. This experiment validated the accuracy of the CNVb marker and proved the authenticity of the CNVb marker-based strategy in distinguishing multiple allele types, even without fully assembled sequences, saving effort compared to traditional SNP/InDel/SSR markers.
Digital fingerprinting map of wheat varieties utilizing CNVb markers
We constructed a comprehensive CNVb fingerprint map consisting of 1599 accessions, by further integrating public resequencing data of 1071 wheat accessions [ 39 , 40 , 41 ] (Additional file 1 : Table S1). Moreover, we created a QR-code-like two-dimensional CNVb markers profile for each accession (Fig. 4 a). The presence of a CNVb-duplication or a CNVb-deletion marker in a variety indicates that this variety contains the duplication or deletion block, respectively. For instance, the profile of Lunxuan987 showed that 276 CNVb markers were detected as present, including the 1RS·1BL translocation marker and perInv-6B marker. In the CNVb fingerprint map, the number of CNVb markers present in each accession ranges from 119 to 322. The genotypes of 199 markers are different between pairwise accessions on average (Fig. 4 b), and there are more than 100 markers with different genotypes for 99.5% of the accession pairs (Fig. 4 b). For example, the sibling cultivars Bima1 and Bima4 present 117 distinct markers (Additional file 2 : Fig. S10). These results indicate a great potential of CNVb markers for discriminating closely related accessions. To further evaluate the accuracy of variety discrimination using CNVb markers, we compared CNVb strategy and germplasm resource-based Identity-By-Descent (gIBD), a previous strategy evaluating the genome-wide similarity that could reflect pedigree relationships in various plant species [ 16 ]. The results showed that the similarity estimated by the CNVb-based strategy is highly correlated with gIBD-based similarity, and the correlation is especially significant for varieties with a close genetic relationship (similarity > 0.4, Pearson’s correlation = 0.85, P < 2.2 × 10 −12 ) (Fig. 4 c). Thus, the result demonstrated the CNVb markers can serve as an effective strategy for the reliable estimation of genetic similarity to distinguish the genetic-similar wheat accessions.

Performance evaluation of CNVb markers in germplasm identification. a The CNVb marker fingerprint of Lunxuan987. CNVb marker fingerprint consists of a QR-code-like two-dimensional matrix, with each cell representing a CNVb marker. All the markers are ordered by chromosomes and are filled into the matrix by rows, from left to right and from top to bottom. Two specific markers were highlighted by arrows with annotated descriptions as interspecific introgression or structural variation. b Spectrum of the number of CNVb markers in each accession and the number of differential CNVb markers in pairwise accessions. c Variety similarity was calculated based on CNVb markers and germplasm resource-based Identity-By-Descent (gIBD) block, respectively. Varieties with similarities calculated by both methods above 0.4 are highlighted in blue in the upper right corner. The upper right corner also displays the regression trend between a variety of similarities calculated based on CNVb markers and gIBD. Vertical and horizontal dashed lines represent variety similarity equal to 0.4, respectively. r , Pearson’s correlation coefficient, P value < 2.2 × 10 −12 . d The accuracy of identifying CNVb fingerprints in accessions at low sequencing coverage. The ulcWGS data are simulated from whole genome sequencing data of 100 accessions randomly selected from the original CNVb marker library construction. e The similarity of pairwise accessions from two batches of 0.05× simulated sequencing datasets, each comprising 100 randomly selected accessions from our dataset. The dashed line represents the threshold (85%) for variety identification. f The similarity of pairwise accessions from two batches of 0.05× sequencing data, each containing 100 accessions not included in the original CNVb marker library. The dashed line represents the similarity threshold (85%) for variety identification
Boost germplasm identification with ultra-low-coverage sequencing
Genetic identification of germplasm resources is crucial for protecting breeders’ rights and promoting its digital management. Reliability tests of CNVb markers among various depths (0.05×, 0.1×, 0.5×, 1.0×, and 1.5×) showed that the minimum recall and precision ratio observed at these reduced depths were above 99.0% and 97.9%, respectively, for each accession (Fig. 4 d), demonstrating the robustness of CNVb fingerprints in ulcWGS. To further examine the power of CNVb fingerprints in distinguishing germplasm under ulcWGS, we selected 100 accessions from the original CNVb marker library and compared the CNVb fingerprints estimated at both the original and downsampled sequencing coverages. The similarity between pairwise accessions exhibited a bimodal distribution with two distinct peaks, which corresponded to the similarity between the same varieties and between different varieties (Additional file 2 : Fig. S11). A similarity of 85% was selected as the threshold for variety differentiation based on the 99% confidence interval of the “distinct variety” distribution to ensure high accuracy in distinguishing varieties. The results showed that more than 99.9% of varieties could be accurately classified when the sequencing depth surpassed 0.05× (Fig. 4 e, Additional file 2 : Fig. S12), verifying the CNVb fingerprint-based germplasm identification strategy at ultra-low sequencing coverage. To assess the generalization ability of this strategy, 100 accessions not among the original accessions used to construct the CNVb marker library were randomly selected and subjected to two rounds of downsampling to 0.05× ulcWGS data, creating two replicate datasets. Pairwise comparisons of these accessions confirmed that the strategy with a threshold of 85% can effectively differentiate accessions, as well as replications of the same accessions (Fig. 4 f). This demonstrates the practicality and accuracy of CNVb markers in ulcWGS for germplasm identification.
WheatCNVb database for exploring and comparing CNVb profiles
To enhance the accessibility of CNVb markers, we developed a database named WheatCNVb ( http://wheat.cau.edu.cn/WheatCNVb/ ), based on the profiling of 1599 hexaploid wheat accessions with 1240 CNVb markers. Generally, the WheatCNVb database offers four main functions. First, the “CNVb profile” function allows users to query the CNVb profile for each accession. Two visualization modes were offered, as a chromosomal profile with colored regions representing the presented markers, and a QR-code-like representation of the digital present-absent status of 1240 CNVb markers (Fig. 5 a). Second, the “CNVb marker info” function provides detailed information on CNVb marker, including the marker ID, location, annotations, and the accession list that harbors this marker (Fig. 5 b). Third, the “Variety compare” function supports the comparison of CNVb fingerprints for any selected pair of accessions, which can intuitively visualize the shared and differential CNVb markers and estimate the similarity based on the CNVb profiles (Fig. 5 c). For example, a pairwise analysis using the WheatCNVb database revealed a 52.6% genetic similarity between Jimai22 and Jimai20 (Fig. 5 c). Additionally, each variety possesses 90 and 80 unique CNVb markers, respectively (Fig. 5 c), confirming their classification as distinct wheat varieties.

Schematic of the WheatCNVb database. a The “CNVb profile” presents an example of the distribution of the CNVb marker and the CNVb fingerprint barcode of Jagger. b The “CNVb marker info” function provides a table including marker ID, location, introgression source, and relevant accessions of each CNVb marker. c The “Variety compare” function shows the CNVb fingerprints in the mode of pairwise comparison, also with the estimated similarity. d The “Geno scan” function allows users to upload a bin-wised read-depth profile, which can be calculated with ultra-low whole genome sequencing data, and generate accession-specific CNVb fingerprint for variety identification and similarity evaluation
Moreover, the “Geno scan” function enables users to analyze customized wheat accessions. Users can perform an ulcWGS to their material, locally prepare the bin-wised read depth file of the accession locally with a pipeline provided on the webpage ( http://wheat.cau.edu.cn/WheatCNVb/tutorial.html ), and upload the file to the WheatCNVb database (Fig. 5 d). The database will facilitate the identification of CNVb markers for the accession, obtaining a CNVb fingerprint that can be compared with varieties stored in the database or other submitted varieties for comprehensive variety identification.
High-throughput, affordable, and rapid detection of DNA-based markers is essential for exploring germplasm diversity and protecting breeders’ rights. However, developing an efficient genotyping tool for crops like wheat, with its huge and complex genome, remains challenging. Despite the abundance of SVs (including CNVs), which are crucial polymorphisms in crops, efforts to develop automated platforms for CNV typing are limited [ 13 ]. In this study, we revealed that the high frequency and polymorphism of large CNV blocks in wheat make CNVb an effective DNA-based marker for efficient variety identification. We generated a comprehensive reference catalog of CNV blocks at the pan-genome level, which captures sequence polymorphisms absent in Chinese Spring and provides sufficient CNVb markers to perform accurate variety identification. Additionally, we addressed the high rates of false positives and negatives in CNVb calling specific to ulcWGS by refining and merging raw CNV blocks. We manually annotated the tagged CNVb markers with known structural variations and beneficial alleles, and we developed an ulcWGS scanning strategy for new candidate varieties, which demonstrated advanced performance in germplasm identification.
Genotyping gene bank collections is a crucial first step in harnessing the untapped biodiversity of wheat genetic resources [ 42 ]. To date, over 560,000 wheat accessions are preserved in near 40 gene banks worldwide [ 11 ]. Despite significant progress in collecting wheat resources, the capacity to identify, integrate, and utilize such extensive germplasm remains markedly insufficient [ 3 , 43 ]. Compared to conventional methods for assessing wheat genetic resources in gene banks, CNVb markers showed multiple aspects of advantages (Additional file 2 : Table S9). First, CNVb markers significantly reduced the cost for genotyping per marker compared to Southern blot-based markers like RFLP and chip-based markers like SNP arrays, while being comparable in cost-effectiveness to SSRs and GBS. Second, CNVb markers support ultra-low-depth high-throughput sequencing and can be fully automated, which is more labor-saving and less equipment-dependent than widely used SSR markers, making them more suitable for large-scale applications. Third, CNVb markers provide very high reliability, comparable to SNP arrays, and better performance than the GBS strategy. Fourth, CNVb markers provide high accuracy in variety identification, capable of distinguishing even closely related accessions (>40% similarity), comparable to genome-wide gIBD analysis using high-coverage whole genome sequencing (Fig. 4 c). Fifth, CNVb markers support capturing larger genomic variations, which provides unique genetic information and is crucial for identifying traits linked to structural variations. This feature is particularly advantageous in polyploid crops like wheat, where large genomic structural variations are prevalent. Thus, CNVb markers represent a low-cost, high-throughput, labor-saving, and highly reliable tool for modern breeding and germplasm management. The current plant variety protection system relies on phenotype-based distinctness, uniformity, and stability assessments, which can be costly, time-consuming, and often limited to a small number of traits influenced by environmental conditions [ 8 ]. Moreover, with the emergence of new breeding technologies that facilitate minor modifications in varieties, yielding specific merits or utilities, the challenge of detecting distinctness between varieties, especially those that are essentially derived, is increasing [ 44 , 45 ]. CNVb marker is cost-efficient, high throughput, and highly accurate, making it a practical alternative to morphological trait and traditional molecular markers. It provides a low-cost, thousand-marker one-time, and rapid technical solution, ideal for establishing an evaluation system for essentially derived varieties.
The initial hybridization of bread wheat involved a limited number of individuals, where the diploid Aegilops tauschii (DD) was hybridized with the tetraploid Triticum turgidum (AABB) to form the allohexaploid Triticum aestivum (AABBDD), resulting in lower genetic diversity compared to its progenitors [ 46 ]. To address this, farmers and early breeders incorporated members from secondary and tertiary gene pools into wheat breeding programs [ 17 , 19 ]. However, the absence of a high-throughput, cost-effective, and precise identification strategy hinders the resolution and utilization of numerous SVs and interspecific introgressions within the wheat genome. Our PCR analysis suggests that CNVb markers can be associated with various types of genomic variations, indicating their potential as effective signals for tracking documented SVs and introgression events.
This study serves as a preliminary exploration for the development of wheat CNVb markers. Our findings suggest that CNVb is the optimal choice for identifying large SVs and introgression within the wheat genome, as well as for variety identification. Considering the current limited availability of resequencing data, and the abundance of whole exome sequencing and microarray data, our future efforts will focus on integrating these data sets to update and expand the CNVb marker collection, which will facilitate the discovery of rare CNVbs. Additionally, associating these markers with phenotype data will help in nominating key CNVb markers to assist in the wheat breeding programs. The CNVb marker identification strategy outlined in this study also shows promise for application in other crops.
Our study introduces a CNVb-based genotyping approach that could enhance the digitalization and management of wheat germplasm resources using ultra-low-coverage sequencing. The CNVb markers, validated by PCR analysis, not only facilitate the discrimination of massive wheat varieties but also link key genetic traits and beneficial alleles. The WheatCNVb platform further supports this approach by providing a dynamic, user-friendly interface for the exploration and comparison of CNVb profiles, embodying a practical tool for breeders and researchers. Overall, the CNVb-based approach promises a low-cost and high-throughput genotyping strategy for enabling digitalized wheat germplasm management and modern breeding with precise and practical decision-making.
Collection and variation calling of wheat resequencing data
A total of 1599 published wheat accessions with whole genome resequencing data [ 16 , 17 , 25 , 26 , 27 , 39 , 40 , 41 ] (Additional file 1 : Table S1) were used in this study. Trimming of raw reads was performed using Trimmomatic, followed by the mapping of high-quality reads to the wheat pan-genome via BWA-MEM [ 47 ]. Bamtools v2.4 [ 48 ] was used to filter read pairs with either abnormal insert sizes (>10,000 bp or =0 bp) or low mapping quality scores (<1). Samtools v1.3 [ 49 ] was then employed to remove any potential PCR duplicate reads.
Construct non-Chinese Spring chromosome at pan-genome level (chrNCP)
To construct a wheat pan-genome, we first collected de novo assembled genomes of 17 wheat varieties [ 15 , 26 , 28 , 31 , 32 , 33 , 34 , 35 ], including the reference assembly of Chinese Spring RefSeq v1 (CS). Excluding CS, the remaining 16 genomes were ranked based on contig N50 length and whether Hi-C sequencing was used for scaffolding (Additional file 1 : Table S2). We identified absent sequences in the CS genome from the 16 varieties using a whole-genome iterative alignment strategy. The alignment process involved trimming raw reads using Trimmomatic, followed by mapping high-quality reads to the wheat pan-genome with BWA-MEM [ 47 ]. Bamtools v2.4 [ 48 ] was used to filter read pairs with abnormal insert sizes (>10,000 bp or =0 bp) or low mapping quality scores (<1). Samtools v1.3 [ 49 ] was employed to remove potential PCR duplicate reads. Starting with the highest-ranked genome Aikang58 genome as the reference, we aligned CS resequencing data to Aikang58, using a 1 Mb sliding window and a read-depth based method to detect sequences absent in CS relative to Aikang58. This procedure was iteratively applied, comparing CS and Aikang58 resequencing data against the second-ranked Fielder genome to identify non-redundant deletion blocks relative to Fielder, and continued through all 16 varieties. Through this methodology, we extracted non-redundant deletion block sequences absent in CS, which were assembled in chromosomal order into “chrNCP” as a supplementary genome sequence to the CS reference. Thus, “chrNCP” combined with the CS genome forms the wheat pan-genome (Additional file 1 : Table S4).
Identification of CNV blocks
The genome was segmented into 100 Kb nonoverlapping windows to calculate the average read depth, utilizing the “coverage” function in bedtools v2.27.1 [ 50 ]. These counts were then normalized by dividing them by the mode of the read depth across the genome. According to the distribution pattern of normalized read counts, which showed a near-normal distribution centered around a value of 1, windows exhibiting normalized read counts below 0.5 or above 1.5 were classified as deletion and duplication windows, respectively. Finally, contiguous deletion and duplication windows were merged to delineate whole-genome CNV blocks.
Development of CNVb markers
To develop CNVb markers from 528 resequenced varieties, the identification of raw CNV blocks was refined through a systematic process structured into three main steps.
Step 1: Filtering of raw CNV blocks. Initially, for CNV blocks aligned to the CS reference, we employed a multinomial hidden Markov model (HMM) using the hmmlearn Python library ( https://pypi.org/project/hmmlearn/ ) to minimize random noise and enhance the clarity of CNV block patterns. This model was configured with parameters set to “n_components=3, n_iter=60, tol=0.001” and optimized via the Baum-Welch iterative re-estimation algorithm through the “fit()” method. The “decode()” method, with “algorithm=viterbi”, was then used to smooth and decode CNV blocks. CNV blocks with a value of (length / 100 Kb + N) ≤ 10 were further filtered out, where “N” indicates the number of accessions containing the CNV block and “length” indicates the length of CNV block. For CNV blocks mapped to the “chrNCP” genome, a similar filtration and refinement were applied, excluding CNV blocks with a value of (length / 1 Mb + N) ≤ 10 or (length / 1 Mb + n) ≤ 10, where “n” indicates the number of accessions without the CNV block.
Step 2: Merging CNV blocks. For CNV blocks within the CS reference regions, redundancy was addressed by merging significantly overlapping blocks ( ρ o ≥ 0.8) and merging linked blocks (those within 5 Mb apart and with ρ link ≥ 0.9). The formulas for ρ o and ρ link are defined as:
where L 1 and L 2 are the lengths of the CNV blocks, L o is the overlapping length, C 1 and C 2 are the counts of accessions carrying each CNV block, and C s is the count of accessions with both CNV blocks. No further processing was needed for already filtered CNV blocks corresponding to the “chrNCP” genome. This merging step resulted in a preliminary CNV marker library, encompassing multiple CNV blocks per marker. Markers identified in both the CS reference and “chrNCP” sequences were assessed for redundancy with a specific focus on their presence or absence across accessions. If the genotype of a marker form “chrNCP” is highly correlated with that of another marker from CS, the marker from the “chrNCP” sequence will be filtered out.
Step 3: Filtering CNVb markers for ulcWGS stability. To ensure the applicability of CNVb markers for ulcWGS data, markers indistinguishable at low sequencing coverage were excluded. CNV blocks were initially genotyped from hcWGS and simulated 0.1× coverage data, with the latter obtained by downsampling hcWGS data. Each accession’s CNV blocks were compared with the preliminary marker library to ascertain the presence or absence of CNVb markers in both hcWGS and 0.1× coverage data. A marker was considered present if at least one CNV block overlapped with it by ≥90% and the length discrepancy between the CNV block and the marker is less than 1 Mb. Markers with inconsistent detections in more than 10 accessions were removed. The refined set of CNVb markers formed the finalized marker collection.
Construction of the low-coverage sequencing test set
To create a test set for ulcWGS, 100 accessions with sequencing depths > 5× were randomly selected (Additional file 1 : Table S5). Their original BAM files were downsampled to depth levels of 0.01×, 0.05×, 0.1×, 0.5×, 1×, and 1.5×, thereby generating simulated ulcWGS data using Samtools v1.3.1 [ 49 ]. CNV blocks were then genotyped for each accession’s ulcWGS data. These identified CNV blocks from each accession were compared with the raw CNVb marker library to ascertain the presence or absence of each CNVb marker in the simulated ulcWGS data.
Identification of CNVb markers using ulcWGS
The pipeline is to first identify the type of CNV blocks and then match these CNV blocks to the corresponding markers to identify which markers are present in each variety (Additional file 1 : Fig. S4). Initially, the ulcWGS data are aligned to the pan-genome to detect raw deletion (copy number = 0) and duplication (copy number ≥ 2) blocks. These blocks are then separately compared with their corresponding CNVb marker set. The presence of a deletion or duplication marker in a variety is determined based on the following criteria, if a deletion or duplication block present in the variety overlaps with a deletion or duplication marker by at least 90% and the difference in length between the block and the marker is less than 100 Kb.
Evaluating lcWGS recall for SNPs, raw CNVb, and CNVb markers
SNPs were detected in all 100 accessions using GATK v3.868’s HaplotypeCaller module in GVCF mode. To assess the recall rates for SNPs, raw CNVb, and CNVb markers identified via low-coverage sequencing, these findings were benchmarked against results from high-coverage sequencing.
PCR analysis
The primer sequences for three marker types were designed based on distinct introgression fragments. Type 1 marker primers were derived from an introgression fragment in the Jagger genome, corresponding to the CNVb-deletion type 1 (CNVb.139, chr2A: 0–24.7 Mb). The forward primer was 5′-TGCATGTCACTACCACGACC-3′, and the reverse primer was 5′-ACAACCCGTTTTCTTCACGG-3′. Type 2 marker primers were selected from an introgression fragment in the Zang1817 genome, corresponding to the CNVb-deletion type 2 (CNVb.142, chr2A: 12.0–21.3 Mb). The forward primer was 5′-TACTTTCGGATTGACAATTATCCTCTTATC-3′, and the reverse primer was 5′-TGGAAAAATGGTCTTACGGTTATATGAAAT-3′. For the type 3 marker, primers were selected from a segment of the CS genome sequence, aligning with the region of CNVb-deletion type 2 (CNVb.142, chr2A: 12.0–21.3 Mb). The forward primer was 5′-GAACTGATTACAAATGAATAGTTGTAGGGA-3′, and the reverse primer was 5′-TTAGTTACACCATGAGTTAGCATCATTTAG-3′. The PCR reaction system was 20 μL, including 10 μL 2× M5 HiPer plus Taq HiFi PCR mix, 1 μL forward primers and 1 μL reverse primers (10 μmol L −1 ), 2 μL template DNA (150 ng μL −1 ), supplemented with ddH 2 O to 20 μL. The PCR conditions were 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 5 min, and finally followed by 72 °C for 5 min.
Calculation of the pairwise similarity
The pairwise similarity between accessions was calculated based on their CNVb fingerprints. The formula for similarity is defined as:
where M S 1 and M S 2 represent the number of markers in the first and second accessions, respectively, and M share denotes the number of markers shared between the two accessions.
Assessing the accuracy of variety identification based on ulcWGS
This study evaluates the accuracy of variety identification using ulcWGS by comparing it with hcWGS. The test set of 100 accessions from ulcWGS is designated as replicate 1, while an identical set of 100 accessions sequenced at a high depth forms replicate 2. CNVb fingerprints are used for pairwise comparisons between the replicates to simulate the variety identification process. The similarity between each pair is calculated, with a threshold of 85% similarity set to determine if the accessions are of the same or different varieties. Power of variety identification is defined as the proportion of correctly identified distinct variety pairs out of the total distinct pairs.
In addition, we randomly selected 100 accessions that were not among accessions used to construct the CNVb marker library and performed downsampling on their original BAM files to 0.05× coverage in two separate batches using Samtools v1.3.1 [ 49 ]. This process generated two sets of 0.05× simulated sequencing data. The CNVb fingerprints from both data sets were then subject to pairwise comparisons, designating the first data set as replicate 1 and the second as replicate 2. We calculated the similarity between the two replicates, setting a similarity threshold of 85% for variety identification. The statistical power (1 − β) was also computed as the standard for evaluating the accuracy of varietal identification.
Availability of data and materials
The raw reads of 1599 previously published resequenced accessions are available under the following NCBI Sequence Read Archive accessions: PRJNA544491 [ 15 ], PRJNA722149 [ 16 ], PRJNA476679 [ 17 ], PRJNA597250 [ 25 ], PRJNA596843 [ 26 ], PRJNA439156, PRJNA663409 [ 27 ], PRJEB48988, PRJEB48738 [ 39 ], and the National Genomics Data Center ( https://bigd.big.ac.cn/gwh ) database under project CRA005878 [ 40 ]. The previously published de novo assembled reference genome sequences are available under the following NCBI Sequence Read Archive accessions: PRJNA544491, PRJEB37938, PRJNA492239, PRJNA528431, PRJEB39558, PRJEB35709 [ 15 ], PRJNA595806 [ 26 ], PRJEB44721 [ 32 ], PRJEB45541 [ 33 ], and PRJEB49351 [ 34 ]. They are also available in the National Genomics Data Center ( https://bigd.big.ac.cn/gwh ) database under accession number GWHANRF00000000 [ 31 ] and the BIG Data Center ( https://bigd.big.ac.cn/ ) under BioProject numbers PRJCA004332 [ 35 ]. The database WheatCNVb is available as open-source code under the MIT License at https://github.com/Niujx98/WheatCNVbDB [ 51 ] and the raw data can be accessed on Zenodo ( https://doi.org/10.5281/zenodo.11403154 ) [ 52 ]. The CNVb detection pipeline and corresponding manual are available as open-source code under the MIT License at https://github.com/Niujx98/WheatCNVbScan [ 53 ] and can be accessed on Zenodo ( https://doi.org/10.5281/zenodo.11401875 ) [ 54 ].
Shiferaw B, Smale M, Braun H-J, Duveiller E, Reynolds M, Muricho G. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Security 2013, 5: 291-317.
Milner SG, Jost M, Taketa S, Mazon ER, Himmelbach A, Oppermann M, Weise S, Knupffer H, Basterrechea M, Konig P, et al. Genebank genomics highlights the diversity of a global barley collection. Nat Genet. 2019;51:319–26.
Article CAS PubMed Google Scholar
Mascher M, Schreiber M, Scholz U, Graner A, Reif JC, Stein N. Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat Genet. 2019;51:1076–81.
Ho SS, Urban AE, Mills RE. Structural variation in the sequencing era. Nat Rev Genet. 2020;21:171–89.
Chiang C, Scott AJ, Davis JR, Tsang EK, Li X, Kim Y, Hadzic T, Damani FN, Ganel L, Consortium GT, et al. The impact of structural variation on human gene expression. Nat Genet 2017, 49: 692-699.
Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 2020;182(145–161): e123.
Google Scholar
Yuan Y, Bayer PE, Batley J, Edwards D. Current status of structural variation studies in plants. Plant Biotechnol J. 2021;19:2153–63.
Article PubMed PubMed Central Google Scholar
Jamali SH, Cockram J, Hickey LT. Insights into deployment of DNA markers in plant variety protection and registration. Theor Appl Genet. 2019;132:1911–29.
Rasheed A, Xia X. From markers to genome-based breeding in wheat. Theor Appl Genet. 2019;132:767–84.
Grover A, Sharma PC. Development and use of molecular markers: past and present. Crit Rev Biotechnol. 2016;36:290–302.
Sansaloni C, Franco J, Santos B, Percival-Alwyn L, Singh S, Petroli C, Campos J, Dreher K, Payne T, Marshall D, et al. Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nat Commun. 2020;11:4572.
Article CAS PubMed PubMed Central Google Scholar
Adhikari L, Raupp J, Wu S, Wilson D, Evers B, Koo DH, Singh N, Friebe B, Poland J. Genetic characterization and curation of diploid A-genome wheat species. Plant Physiol. 2022;188:2101–14.
Rasheed A, Hao Y, Xia X, Khan A, Xu Y, Varshney RK, He Z. Crop breeding chips and genotyping platforms: progress, challenges, and perspectives. Mol Plant. 2017;10:1047–64.
Dubcovsky JD, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 2007, 316: 5.
Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D, et al. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588:277–83.
Yang Z, Wang Z, Wang W, Xie X, Chai L, Wang X, Feng X, Li J, Peng H, Su Z, et al. ggComp enables dissection of germplasm resources and construction of a multiscale germplasm network in wheat. Plant Physiol. 2022;188:1950–65.
Cheng H, Liu J, Wen J, Nie X, Xu L, Chen N, Li Z, Wang Q, Zheng Z, Li M, et al. Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol. 2019;20:136.
Wang Z, Wang W, Xie X, Wang Y, Yang Z, Peng H, Xin M, Yao Y, Hu Z, Liu J, et al. Dispersed emergence and protracted domestication of polyploid wheat uncovered by mosaic ancestral haploblock inference. Nat Commun. 2022;13:3891.
Przewieslik-Allen AM, Wilkinson PA, Burridge AJ, Winfield MO, Dai X, Beaumont M, King J, Yang CY, Griffiths S, Wingen LU, et al. The role of gene flow and chromosomal instability in shaping the bread wheat genome. Nat Plants. 2021;7:172–83.
Huang X, Zhu M, Zhuang L, Zhang S, Wang J, Chen X, Wang D, Chen J, Bao Y, Guo J, et al. Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor Appl Genet. 2018;131:1967–86.
Wu N, Lei Y, Pei D, Wu H, Liu X, Fang J, Guo J, Wang C, Guo J, Zhang J, et al. Predominant wheat-alien chromosome translocations in newly developed wheat of China. Molecular Breeding 2021, 41 .
Kosugi S, Momozawa Y, Liu X, Terao C, Kubo M, Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117.
Mahmoud M, Gobet N, Cruz-Davalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. Structural variant calling: the long and the short of it. Genome Biol. 2019;20:246.
Keilwagen J, Lehnert H, Berner T, Badaeva E, Himmelbach A, Borner A, Kilian B. Detecting major introgressions in wheat and their putative origins using coverage analysis. Sci Rep. 1908;2022:12.
Hao C, Jiao C, Hou J, Li T, Liu H, Wang Y, Zheng J, Liu H, Bi Z, Xu F, et al. Resequencing of 145 landmark cultivars reveals asymmetric sub-genome selection and strong founder genotype effects on wheat breeding in China. Mol Plant. 2020;13:1733–51.
Guo W, Xin M, Wang Z, Yao Y, Hu Z, Song W, Yu K, Chen Y, Wang X, Guan P, et al. Origin and adaptation to high altitude of Tibetan semi-wild wheat. Nat Commun. 2020;11:5085.
Zhou Y, Zhao X, Li Y, Xu J, Bi A, Kang L, Xu D, Chen H, Wang Y, Wang YG, et al. Triticum population sequencing provides insights into wheat adaptation. Nat Genet. 2020;52:1412–22.
International Wheat Genome Sequencing Consortium et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361(6403):eaar7191.
Zhang X, Zhu Y, Kremling KAG, et al. Genome-wide analysis of deletions in maize population reveals abundant genetic diversity and functional impact. Theoretical and Applied Genetics. 2021;135(1):273–90.
Article PubMed Google Scholar
Bai Z, Chen J, Liao Y, Wang M, Liu R, Ge S, Wing RA, Chen M. The impact and origin of copy number variations in the Oryza species. BMC Genomics. 2016;17:261.
Jia J, Zhao G, Li D, Wang K, Kong C, Deng P, Yan X, Zhang X, Lu Z, Xu S, et al. Genome resources for the elite bread wheat cultivar Aikang 58 and mining of elite homeologous haplotypes for accelerating wheat improvement. Mol Plant. 2023;16:1893–910.
Sato K, Abe F, Mascher M, Haberer G, Gundlach H, Spannagl M, Shirasawa K, Isobe S. Chromosome-scale genome assembly of the transformation-amenable common wheat cultivar ‘Fielder’. DNA Res 2021, 28 .
Athiyannan N, Abrouk M, Boshoff WHP, Cauet S, Rodde N, Kudrna D, Mohammed N, Bettgenhaeuser J, Botha KS, Derman SS, et al. Long-read genome sequencing of bread wheat facilitates disease resistance gene cloning. Nat Genet. 2022;54:227–31.
Aury JM, Engelen S, Istace B, Monat C, Lasserre-Zuber P, Belser C, Cruaud C, Rimbert H, Leroy P, Arribat S, et al. Long-read and chromosome-scale assembly of the hexaploid wheat genome achieves high resolution for research and breeding. Gigascience 2022, 11 .
Shi X, Cui F, Han X, He Y, Zhao L, Zhang N, Zhang H, Zhu H, Liu Z, Ma B, et al. Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar Kenong 9204. Mol Plant. 2022;15:1440–56.
Song L, Liu J, Cao B, Liu B, Zhang X, Chen Z, Dong C, Liu X, Zhang Z, Wang W, et al. Reducing brassinosteroid signaling enhances grain yield in semi-dwarf wheat. Nature. 2023;617:118–24.
Delorean E, Gao L, Lopez JFC, Open Wild Wheat C, Wulff BBH, Ibba MI, Poland J. High molecular weight glutenin gene diversity in Aegilops tauschii demonstrates unique origin of superior wheat quality. Commun Biol 2021, 4: 1242.
Wang X, Song R, An Y, et al. Allelic variation and genetic diversity of HMW glutenin subunits in Chinese wheat landraces and commercial cultivars. Breeding Science. 2022;72(2):169–80.
Schulthess AW, Kale SM, Liu F, Zhao Y, Philipp N, Rembe M, Jiang Y, Beukert U, Serfling A, Himmelbach A, et al. Genomics-informed prebreeding unlocks the diversity in genebanks for wheat improvement. Nat Genet. 2022;54:1544–52.
Niu J, Ma S, Zheng S, Zhang C, Lu Y, Si Y, Tian S, Shi X, Liu X, Naeem MK, et al. Whole-genome sequencing of diverse wheat accessions uncovers genetic changes during modern breeding in China and the United States. Plant Cell. 2023;35:4199–216.
Wang W, Wang Z, Li X, et al. SnpHub: an easy-to-set-up web server framework for exploring large-scale genomic variation data in the post-genomic era with applications in wheat. GigaScience 2020, 9(6).
Longin CF, Reif JC. Redesigning the exploitation of wheat genetic resources. Trends Plant Sci. 2014;19:631–6.
Bohra A, Kilian B, Sivasankar S, Caccamo M, Mba C, McCouch SR, Varshney RK. Reap the crop wild relatives for breeding future crops. Trends Biotechnol. 2022;40:412–31.
Noli E, Teriaca MS, Conti S, Gill K. Criteria for the definition of similarity thresholds for identifying essentially derived varieties. Plant Breeding. 2013;132:525–31.
Article Google Scholar
Yu JK, Chung YS. Plant variety protection: current practices and insights. Genes (Basel) 2021, 12 .
Pont C, Leroy T, Seidel M, Tondelli A, Duchemin W, Armisen D, Lang D, Bustos-Korts D, Goue N, Balfourier F, et al. Tracing the ancestry of modern bread wheats. Nat Genet. 2019;51:905–11.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Barnett DW, Garrison EK, Quinlan AR, Stromberg MP, Marth GT. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–2.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25: 2078-2079.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Niu J, Wang W, Wang Z, Chen Z, Zhang X, Qin Z, Miao L, Yang Z, Xie C, Xin M, et al. Tagging large CNV blocks in wheat boosts digitalization of germplasm resources by ultra-low-coverage sequencing. Github. https://github.com/Niujx98/WheatCNVbDB . 2024.
Niu J, Wang W, Wang Z, Chen Z, Zhang X, Qin Z, Miao L, Yang Z, Xie C, Xin M, et al. Tagging large CNV blocks in wheat boosts digitalization of germplasm resources by ultra-low-coverage sequencing. 2024. Zenodo. https://doi.org/10.5281/zenodo.11403154.Accessed31May .
Niu J, Wang W, Wang Z, Chen Z, Zhang X, Qin Z, Miao L, Yang Z, Xie C, Xin M, et al. Tagging large CNV blocks in wheat boosts digitalization of germplasm resources by ultra-low-coverage sequencing. Github. https://github.com/Niujx98/WheatCNVbScan . 2024.
Niu J, Wang W, Wang Z, Chen Z, Zhang X, Qin Z, Miao L, Yang Z, Xie C, Xin M, et al. Tagging large CNV blocks in wheat boosts digitalization of germplasm resources by ultra-low-coverage sequencing. 2024. Zenodo. https://doi.org/10.5281/zenodo.11401875.Accessed31May .
Download references
Acknowledgements
We thank Prof. Hongqing Ling for providing the Illumina sequencing data. We thank Prof. Zengjun Qi for the discussion on interpreting the data.
Review history
The review history is available as Additional file 3 .
This research was supported by the National Key Research and Development Program of China (grant No. 2020YFE0202300), the National Natural Science Foundation of China (grant No. 32322059), the Pinduoduo-China Agricultural University Research Fund (grant No. PC2023B01016), and the 2115 Talent Development Program of China Agricultural University. This work was supported by the National Postdoctoral Program for Innovative Talents (BX20230414). This work was also supported by the High-performance Computing Platform of China Agricultural University.
Author information
Authors and affiliations.
Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing, 100193, China
Jianxia Niu, Wenxi Wang, Zihao Wang, Zhe Chen, Xiaoyu Zhang, Zhen Qin, Lingfeng Miao, Zhengzhao Yang, Chaojie Xie, Mingming Xin, Huiru Peng, Yingyin Yao, Jie Liu, Zhongfu Ni, Qixin Sun & Weilong Guo
Sanya Institute of China Agricultural University, Sanya, 572025, China
Jianxia Niu & Zihao Wang
You can also search for this author in PubMed Google Scholar
Contributions
W.G. and Q.S. conceived the idea, coordinated the project, and finalized the manuscript. J.N., W.W., and Z.W. developed algorithms, performed data analysis, and drafted the manuscript. Z.C. performed experiments. X.Z. constructed the pan-genome. Z.Q., Y.Z., C.X., H.P., Y.Y., J.L., and Z.N. interpreted the data. W.G., Q.S., Z.C., Z.Q., Z.Y., and L.M. revised the manuscript. All the authors read and approved the manuscript.
Corresponding authors
Correspondence to Qixin Sun or Weilong Guo .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Competing interests
China Agricultural University has filed a patent application for developing and applications of CNVb markers in wheat.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file1 (xlsx 233 kb), additional file2 (pdf 2597 kb), additional file3 (docx 1516 kb), rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Niu, J., Wang, W., Wang, Z. et al. Tagging large CNV blocks in wheat boosts digitalization of germplasm resources by ultra-low-coverage sequencing. Genome Biol 25 , 171 (2024). https://doi.org/10.1186/s13059-024-03315-6
Download citation
Received : 31 January 2024
Accepted : 18 June 2024
Published : 01 July 2024
DOI : https://doi.org/10.1186/s13059-024-03315-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Copy number variation
- Digitalized fingerprinting
- Introgression
- Low-coverage sequencing
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]
If you're seeing this message, it means we're having trouble loading external resources on our website.
If you're behind a web filter, please make sure that the domains *.kastatic.org and *.kasandbox.org are unblocked.
To log in and use all the features of Khan Academy, please enable JavaScript in your browser.
Middle school biology
Middle school earth and space science, middle school physics, high school biology, high school chemistry, high school physics, ap®︎/college biology, ap®︎/college chemistry, ap®︎/college environmental science, ap®︎/college physics 1, ap®︎/college physics 2, organic chemistry, health and medicine, cosmology and astronomy, electrical engineering, high school biology (tx teks), high school chemistry (tx teks), high school physics (tx teks), biology archive, chemistry archive, physics archive.

IMAGES
VIDEO
COMMENTS
Explore the biology library for legacy biology content, not updated with new courses. Learn about the science of biology, water and life, elements and atoms, macromolecules, cells, genetics, evolution, ecology, and more.
Learn Biology Online. Biology Online is the world's most comprehensive database of Biology terms and topics. Since 2001 it has been the resource of choice for professors, students, and professionals needing answers to Biology questions. Search over 75,000+ terms, news, insights, discoveries, and trends in Biology below: Featured Terms. All Terms.
iBiology offers over 500 biology short films, talks and animations for higher education and professional development. Explore topics such as microbes, CRISPR, Nobel Prize winners, mentoring and more.
XBio is a website that teaches biology through stories of great discoveries by the world's top scientists. It offers unique learning resources, such as videos, slides, papers, and activities, that emphasize evidence-based reasoning and social equity in science education.
Learn the basics of biology with videos, articles, and exercises from Khan Academy. Explore topics such as the scientific method, water and life, and the structure of living things.
Biology overview. Biology at Khan Academy explores life in all its forms, from humans to bacteria and how living things convert energy, grow, and reproduce. The course covers key biological molecules, the role of DNA, and the complexity of cells. It also touches on the intriguing world of viruses and how life continues to change and adapt ...
Sensory Secrets of Penis and Clitoris Unlocked after More Than 150 Years. Mysterious nerve structures called Krause corpuscles respond to specific low-frequency vibrations, scientists finally ...
Welcome to the Biology Library. This Living Library is a principal hub of the LibreTexts project, which is a multi-institutional collaborative venture to develop the next generation of open-access texts to improve postsecondary education at all levels of higher learning. The LibreTexts approach is highly collaborative where an Open Access ...
List of biology websites. This is an annotated list of biological websites, including only notable websites dealing with biology generally and those with a more specific focus. Ask A Biologist - has been hosted by ASU School of Life Sciences since 1997. The website contains a large collection of free content, including images, stories, games ...
HHMI BioInteractive offers free online resources for teaching and learning biology, such as interactive modules, videos, animations, and case studies. Explore topics from genetics to ecology, and join the online community of verified educators.
Biology is the study of living things. It is broken down into many fields, reflecting the complexity of life from the atoms and molecules of biochemistry to the interactions of millions of organisms in ecology. This biology dictionary is here to help you learn about all sorts of biology terms, principles, and life forms.
Biology 2 Curriculum and Course Map Materials. Mitosis and Cancer - Data Analysis on Carcinogenesis. Reinforcement: Photosynthesis. Biology 1 Final Exam Review Guide (Bee Book) Resources for biology students that include worksheets, labs, and student activities. Find everything you need for your biology lessons here!!
BioExplorer shares breaking science news and articles on a variety of topics from the leading universities and research institutions around the globe. It is your ultimate guide to Biological Web Resources ranging from DNA to Plants, Animals, and everything in-between. This site was originally developed for the researchers involved in biological ...
The Biology Notes is an educational niche website related to biology (microbiology, biotechnology, biochemistry, zoology, botany, cell biology, genetics, molecular biology, etc.) and different other branches of biology.
BioMan Biology is the fun place to learn Biology! Here you will find learning games, review games, virtual labs and quizzes that will help you to learn about cells, ecology, genetics, physiology, and much more! Note: If you are a teacher, please check out the teacher section for ways to use the site to increase student engagement and learning.
Founded in 2002 by Nobel Laureate Carl Wieman, the PhET Interactive Simulations project at the University of Colorado Boulder creates free interactive math and science simulations. PhET sims are based on extensive education <a {{0}}>research</a> and engage students through an intuitive, game-like environment where students learn through exploration and discovery.
Welcome to AP®︎/College Biology! In this course you'll explore the amazing world of biology through topics like biochemistry, cell biology, genetics, evolution, and ecology. To make sure you're prepared with the fundamentals, we recommend completing high school biology before diving into AP Biology.
Best Science Websites for Teaching Chemistry Ward's Science featuring Ward's World. Check out Ward's World, a new destination that offers middle schoolers and their teachers free classroom activities, how-to videos, tips, tricks, and resources that make science easier—and more fun! Find chemistry, biology, physics, and earth science.
Answer: The top 5 most popular Biology websites in the world in May 2024 are: 1. nature.com. 2. oup.com. 3. frontiersin.org. 4. biomedcentral.com. 5. doodle.com. The complete Biology websites ranking list: Click here for free access to the top Biology websites in the world, ranked by traffic and engagement.
Unlock Engaging Biology Lessons with Comprehensive Resources! Save precious planning time and elevate your classroom experience with our all-in-one Biology teaching package. This resource includes meticulously crafted lesson PowerPoints, engaging worksheets, and multiple practice question PowerPoints complete with model answers. ...
Laboratory-made plasmids, a workhorse of modern biology, have problems. Researchers performed a systematic assessment of the circular DNA structures by analysing more than 2,500 plasmids produced ...
Marie E. Burns, a professor in the Departments of Ophthalmology and Vision Science, and Cell Biology and Human Anatomy in the School of Medicine, and a core faculty member at the Center for Neuroscience (CNS), has been appointed the CNS interim director, effective July 1, 2024.. Burns succeeds A. Kimberley McAllister, who has served as the center's director since 2016 and who is departing UC ...
Project Description The successful applicants will conduct a two-year field research project to evaluate the distribution, population status, and habitat requirements for crayfish species of greatest conservation need (SGCN) in the Pearl River drainage basin in Louisiana.
B lymphocytes (B cells) Professional antigen presenting cells (APC) and MHC II complexes. Helper T cells. Cytotoxic T cells and MHC I complexes. Review of B cells, CD4+ T cells and CD8+ T cells. Inflammatory response. This unit is part of the Biology library. Browse videos, articles, and exercises by topic.
By converting our sims to HTML5, we make them seamlessly available across platforms and devices. Whether you have laptops, iPads, chromebooks, or BYOD, your favorite PhET sims are always right at your fingertips.Become part of our mission today, and transform the learning experiences of students everywhere!
Molecular Biology and Evolution | 41 | 7 | July 2024. The sacred coca tree, a keystone crop for Indigenous Andean and Amazonian communities for over 8,000 years, has recently become associated with armed conflict and deforestation due to demand for the alkaloid cocaine.
The field of synthetic biology, the science of manipulating biology, has a lot of "cooks in the kitchen," which has both helped it flourish and made it unusually difficult to create a cohesive ...
Marcus Feldman, the Burnet C. and Mildred Finley Wohlford Professor and professor in the Department of Biology, has been selected as the 2024 winner of the Society of Molecular Biology and Evolution's Lifetime Achievement Award. This honor recognizes Feldman's outstanding research during his career and contributions to the field of molecular biology and evolution.
The massive structural variations and frequent introgression highly contribute to the genetic diversity of wheat, while the huge and complex genome of polyploid wheat hinders efficient genotyping of abundant varieties towards accurate identification, management, and exploitation of germplasm resources. We develop a novel workflow that identifies 1240 high-quality large copy number variation ...
Learn science topics from biology, chemistry, physics, and more with hundreds of free online courses and videos. Explore middle school, high school, AP®, and college-level content with interactive exercises and quizzes.