share this!
June 4, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source

Scientists push single-molecule DNA sequencing to the next level
by Gladstone Institutes
In recent years, technologies that allow scientists to study a person's DNA at single-molecule resolution have vastly expanded our knowledge of the human genome, the microbiome, and the genetic basis of disease. With such a detailed view of DNA, it's possible to see genetic variants and structural details that were simply undetectable with earlier sequencing technologies.
However, today's gold-standard methods for single-molecule analysis typically require at least 150,000 human cells —containing millions of individual DNA molecules. That means researchers can't apply these tools when just a few thousand cells are available, such as in many tumor biopsies, limiting the scientific and clinical potential of these technologies.
Now, researchers at Gladstone Institutes have developed two new tools for single-molecule analysis that slash the amount of DNA needed by 90 to 95%. Their work, published in the journal Nature Genetics , shows how these tools could allow scientists to address biological questions they were previously unable to answer.
"We've been working toward creating these methods for a very long time," says Vijay Ramani, Ph.D., assistant investigator at Gladstone and senior author of the study. "We're really excited to see what discoveries will now be possible."
'Tagging' DNA for a clearer view
The first of the new tools, known as "single-molecule real time sequencing by tagmentation," or SMRT-Tag, extends the established protocols for simultaneously mapping the sequence of bases in a long DNA fragment and locations of chemical structures called methyl groups , which lie along the length of the DNA. Methyl groups play a key role in gene expression , making them essential for understanding disease, so it's important to see how they're configured on DNA.
"When we have very little DNA to work with, we can't just make more copies of the DNA and apply our usual protocols," Ramani says. "Making copies would strip away these methylation patterns and introduce other errors."
Instead, his team adapted an approach called "tagmentation," which repurposes the bacterial protein Tn5 to simultaneously cut DNA molecules into more manageable fragments and tag them with chemical components necessary for further analysis.
Tagmentation is already used for sequencing short fragments of DNA when only small amounts of DNA are available—but only limited information can be gleaned from short fragments.
The challenge for Ramani's team was to get the biochemistry of tagmentation just right for breaking up small amounts of DNA into long chunks of about 3,000 to 5,000 base pairs. Their method "tags" the ends of each fragment with hairpin-shaped structures, creating long loops of DNA that can be read reliably by sequencer machinery.
"It was quite a heroic effort by the staff and students in my lab," Ramani says. "We had to test different versions of Tn5 and nearly 100 different conditions with different buffers, enzymes, and temperatures. When you're working with such small amounts of DNA, any issue that causes any DNA loss is that much more of a problem."
Actionable data from small samples
Once they optimized SMRT-Tag, the team demonstrated that it performs as well as established protocols but using far lower amounts of DNA—about the amount found in as few as 10,000 cells.
"Using gold-standard single-molecule sequencing machinery, no one has ever sequenced such a small amount of DNA to the coverage we've now achieved," Ramani says.
Next, his team combined SMRT-Tag with a method they previously developed called SAMOSA, short for "single-molecule adenine methylated oligonucleosome sequencing assay."
SAMOSA reveals additional methylation patterns that reflect chromatin accessibility—or, how easily gene expression machinery can access different stretches of DNA.
Now, with the new SAMOSA-Tag tool, the researchers were able to assess chromatin accessibility with much less DNA than previously needed. To demonstrate, they applied it to prostate cancer cells—some from a patient's initial tumor and some from a tumor that had spread to a different location in the body—that had been transplanted and grown in mice. The method revealed differences in chromatin accessibility that hint at possible key drivers of cancer metastasis.
"This is just one example of how our tools could be applied to clinically relevant samples in cancer and other diseases where DNA is in short supply," says Siva Kasinathan, MD, Ph.D., who co-led the study with Ramani. "We think there's some low-hanging fruit there that could unlock some new biology, which could be important for helping patients down the line."
Kasinathan, a clinical fellow at Lucille Packard Children's Hospital at Stanford University, is a visiting scientist at Gladstone and a longtime collaborator with Ramani.
Ramani's team is now optimizing SMRT-Tag and SAMOSA-Tag to work with even smaller amounts of DNA. They also continue to share and regularly update their protocols online, inviting feedback and collaboration from other researchers. "The community and people involved are really important in the story of this work," Ramani says.
In particular, he highlights his work with Kasinathan, who he met while they were in graduate school together at University of Washington. Together, they conceptualized the study. "It's been so meaningful to work with one of my closest friends to publish what we think will be very impactful work for human health."
Journal information: Nature Genetics
Provided by Gladstone Institutes
Explore further
Feedback to editors

Rate of global warming caused by humans is at an all-time high, say scientists
2 hours ago

Giant viruses discovered on Greenland ice sheet could reduce ice melt
4 hours ago

New study confirms presence of benzene in natural gas and potential for undetectable indoor leaks
5 hours ago
Chasing down a cellular 'short circuit' sheds light on how certain diseases begin

Remarkable new plant species steals nutrients from underground fungi
7 hours ago

Others' words, not firsthand experience, shape scientific and religious belief formation, study finds
8 hours ago

Earliest cattle herds in northern Europe found in the Netherlands

Real-time flood risk visualization via server-based mixed reality enhances accessibility and public safety

Rocky shores of Pacific Northwest show low resilience to changes in climate
9 hours ago
Researchers discover disordered clock protein that sheds new light on circadian rhythms
Relevant physicsforums posts, a dna animation.
May 29, 2024
Probability, genetic disorder related
May 28, 2024
Looking For Today's DNA Knowledge
May 27, 2024
Covid Vaccines Reducing Infections
Human sperm, egg cells mass-generated using ips, and now, here comes covid-19 version ba.2, ba.4, ba.5,....
May 25, 2024
More from Biology and Medical
Related Stories

Developing a machine learning model to explore DNA methylation
Apr 12, 2024

Sequencing multiple RNA base modifications simultaneously: A new era of RNA biology
Sep 29, 2021

Novel single-cell nanopore sequencing tool accelerates analysis of tumor cells
Aug 16, 2023

New method boosts the study of regulation of gene activity
Jul 1, 2022

DNA sequencing method can detect where and how small molecule drugs interact with their targets
Jan 23, 2023

New technique adds to the expanding suite of TAPS capabilities
Sep 20, 2022
Recommended for you

New method for safe and efficient cell transfection developed by researchers
Molecular stop signal identified: The surveillance system of cell division
10 hours ago
Study identifies fungus that breaks down ocean plastic
Jun 3, 2024

Scientists develop new method to match genes to their molecular 'switches'
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Phys.org in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
MIT Technology Review
- Newsletters
The first gene-editing treatment: 10 Breakthrough Technologies 2024
Sickle-cell disease is the first illness to be beaten by CRISPR, but the new treatment comes with an expected price tag of $2 to $3 million.
- Antonio Regalado archive page

CRISPR Therapeutics, Editas Medicine, Precision BioSciences, Vertex Pharmaceuticals
The first gene-editing cure has arrived. Grateful patients are calling it “life changing.”
It was only 11 years ago that scientists first developed the potent DNA-snipping technology called CRISPR. Now they’ve brought CRISPR out of the lab and into real medicine with a treatment that cures the symptoms of sickle-cell disease.
Sickle-cell is caused by inheriting two bad copies of one of the genes that make hemoglobin. Symptoms include bouts of intense pain, and life expectancy with the disease is just 53 years. It affects 1 in 4,000 people in the US, nearly all of them African-American.
So how did this disease become CRISPR’s first success ? A fortuitous fact of biology is part of the answer. Our bodies harbor another way to make hemoglobin that turns off when we’re born. Researchers found that a simple DNA edit to cells from the bone marrow could turn it back on.
Many CRISPR treatments are in trials, but in 2022, Vertex Pharmaceuticals, based in Boston, was first to bring one to regulators for approval. That treatment was for sickle-cell. After their bone marrow was edited, nearly all the patients who volunteered in the trial were pain free.
Good news. But the expected price tag of the gene-editing treatment is $2 to $3 million. And Vertex has no immediate plans to offer it in Africa—where sickle-cell disease is most common, and where it still kills children.
The company says this is because the treatment regimen is so complex. It involves a hospital stay; doctors remove the bone marrow, edit the cells, and then transplant them back. In countries that still struggle to cover basic health needs, the procedure remains too demanding. So simpler, cheaper ways to deliver CRISPR could come next.
Biotechnology and health
Google helped make an exquisitely detailed map of a tiny piece of the human brain.
A small brain sample was sliced into 5,000 pieces, and machine learning helped stitch it back together.
- Cassandra Willyard archive page
That viral video showing a head transplant is a fake. But it might be real someday.
BrainBridge is best understood as the first public billboard for a hugely controversial scheme to defeat death.
The effort to make a breakthrough cancer therapy cheaper
CAR-T cells could revolutionize the treatment of a wide variety of diseases, if only we can make them cheaper.
Beyond Neuralink: Meet the other companies developing brain-computer interfaces
Companies like Synchron, Paradromics, and Precision Neuroscience are also racing to develop brain implants
Stay connected
Get the latest updates from mit technology review.
Discover special offers, top stories, upcoming events, and more.
Thank you for submitting your email!
It looks like something went wrong.
We’re having trouble saving your preferences. Try refreshing this page and updating them one more time. If you continue to get this message, reach out to us at [email protected] with a list of newsletters you’d like to receive.
Featured Topics
Featured series.
A series of random questions answered by Harvard experts.
Explore the Gazette
Read the latest.

Bringing back a long extinct bird

‘The scientist is not in the business of following instructions.’

Glimpse of next-generation internet
“Using twin prime editing is like using two word processors at the same time to simultaneously write different parts of the same paragraph,” explains Harvard Professor David Liu, the paper’s senior author.
Stephanie Mitchell/Harvard file photo
Twin gene-editing system gives twice the efficiency
Yahya Chaudhry
Harvard Correspondent
New technique will allow programmable manipulation of large DNA segments
A team of researchers led by Harvard and Broad Institute scientists has developed twin prime editing, a new, CRISPR-based gene-editing strategy that enables manipulation of gene-sized chunks of DNA in human cells without cutting the DNA double helix.
Because it can make larger edits than previously possible, the new technique could make it possible to study and treat genetic diseases arising from loss of gene function or complex structural mutations, such as hemophilia or Hunter syndrome.
In the paper , published in the journal Nature Biotechnology, the researchers detail how they developed twin prime editing (PE), which uses a prime editor protein and two prime editing guide RNAs (pegRNAs) for the programmable replacement or excision of DNA sequences at endogenous human genomic sites without requiring double-stranded DNA cuts.
When combined with a site-specific recombinase, twin PE allowed integration of gene-sized DNA of more than 5,000 base pairs into the human genome at sites chosen by the researchers. Because large structural variants are found in many human disease-causing alleles, this technique may help treat genetic diseases. The study used twin PE plus recombinase to edit genes linked to Hunter syndrome, phenylketonuria (PKU), Duchenne muscular dystrophy, and hemophilia.
“A major remaining challenge in mammalian cell gene editing is our inability to make targeted gene-sized insertions at sites of our choosing,” said David Liu, the paper’s senior author, Thomas Dudley Cabot Professor of the Natural Sciences, and a core faculty member of the Broad Institute. “Such a capability could advance gene therapy by enabling genes to be restored in their native sequence locations, without increased risk of cancer from semi-random or uncontrolled integration at other locations in the genome.”
This work was performed by members of Liu’s lab, including former postdoctoral fellow Andrew Anzalone, former graduate student Chris Podracky, postdoctoral fellow Xin (Daniel) Gao, and current graduate student Andrew Nelson.
This new method is designed to overcome some of the limitations of existing gene-editing techniques, and build on the strengths of prime editing.
In 2016 the Liu group developed base editing, an efficient and precise genome-editing method that functions like a genetic pencil, chemically rewriting one DNA base to another without completely breaking the DNA backbone. The Liu group developed two classes of base editors that can correct four of the most common kinds of single-letter mutations, which collectively account for about 30 percent of known disease-associated human genetic errors. Three years later, the group developed prime editing, in which a reverse transcriptase directly copies edited DNA sequences into a specified target site from an extended guide RNA, again without requiring double-stranded DNA breaks. Like a genetic word processor, prime editing lets researchers search for one DNA segment and swap it for another.
Now, twin PE lets researchers search and replace larger DNA segments, again without breaking double-stranded DNA.
“Using twin prime editing is like using two word processors at the same time to simultaneously write different parts of the same paragraph,” said Liu, who is also a Howard Hughes Medical Institute Investigator. “In the process, you can create that paragraph more efficiently.”
The researchers developed twin PE by using simultaneous prime edits to create both strands of the edited DNA. By itself, twin PE enables efficient deletions, insertions, and substitutions of hundreds of base pairs, achieving larger edits than simple prime editing. Seeking the ability to make even larger edits of up to thousands of base pairs — a size large enough to include many whole genes — the team installed Bxb1 recombinase “landing sites” at targets site in human cells. The recombinase then mediated the insertion of large DNA cargo sequences into the landing sites.
The team tested twin PE plus Bxb1 recombinase by inserting large DNA into a variety of target sites in the human genome, including at “safe harbor loci” thought to be ideal for gene therapy because inserting genes there has been found to not induce cancer or other apparent toxicities.
The researchers also applied twin PE-mediated deletions to target DMD. Pathogenic DMD alleles, which cause Duchenne muscular dystrophy, commonly contain large deletions in exonic regions that result in frame-shifted transcripts. These experiments show that twin PE can generate large deletions at therapeutically relevant loci in humans with far fewer potentially harmful byproducts than paired Cas9 nuclease strategies.
The team also used twin PE and a recombinase to install a pathogenic 40,000 bp inversion that causes Hunter syndrome. Their success suggests that the combination might eventually work as a therapeutic strategy for correcting other large or complex pathogenic gene variants.
Going forward, Liu is optimistic that other recombinases can work effectively with twin PE.
“We hope to apply twin PE to some basic science and some therapeutic questions, while continuing to develop this strategy to increase the efficiency of large gene size integrations,” Liu said.
Podracky, who joined the lab in 2015, said he hopes that the twin PE plus recombinase technique will both be studied and be an inspiration for scientists across the country.
“We could see other academic labs take the technology and put it put it to good use,” Podracky said. “If this enables [scientists] to realize their gene-editing ideas more readily, that would be a success.”
This study was supported by the Merkin Institute of Transformative Technologies in Healthcare, the National Institutes of Health, and the Howard Hughes Medical Institute.
Share this article
You might like.
Scientists sequence complete genome of bush moa, offering insights into its natural history, possible clues to evolution of flightless birds

George Whitesides became a giant of chemistry by keeping it simple
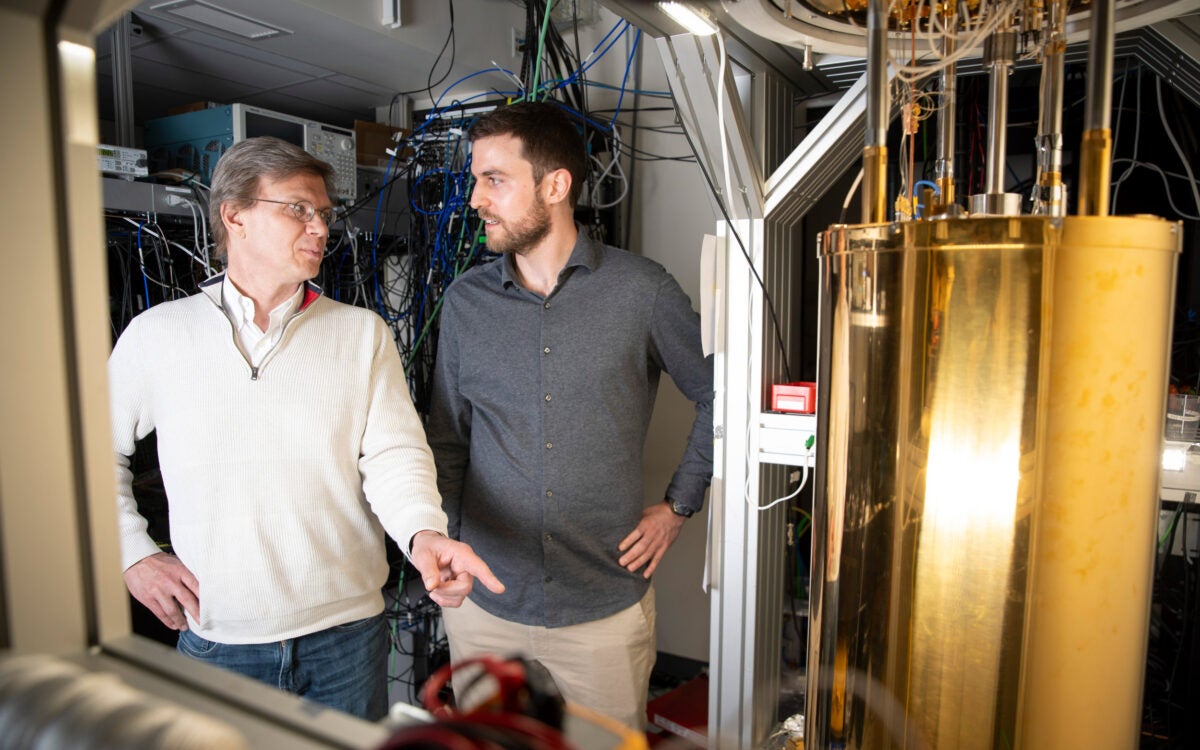
Physicists demo first metro-area quantum computer network in Boston
When should Harvard speak out?
Institutional Voice Working Group provides a roadmap in new report
Women who follow Mediterranean diet live longer
Large study shows benefits against cancer, cardiovascular mortality, also identifies likely biological drivers of better health
Had a bad experience meditating? You're not alone.
Altered states of consciousness through yoga, mindfulness more common than thought and mostly beneficial, study finds — though clinicians ill-equipped to help those who struggle
Along with Stanford news and stories, show me:
- Student information
- Faculty/Staff information
We want to provide announcements, events, leadership messages and resources that are relevant to you. Your selection is stored in a browser cookie which you can remove at any time using “Clear all personalization” below.
A new, nano-scale look at how the SARS-CoV-2 virus replicates in cells may offer greater precision in drug development, a Stanford University team reports in Nature Communications . Using advanced microscopy techniques, the researchers produced what might be some of the most crisp images available of the virus’s RNA and replication structures, which they witnessed form spherical shapes around the nucleus of the infected cell.
“We have not seen COVID infecting cells at this high resolution and known what we are looking at before,” said Stanley Qi , Stanford associate professor of bioengineering in the Schools of Engineering and of Medicine and co-senior author of the paper. “Being able to know what you are looking at with this high resolution over time is fundamentally helpful to virology and future virus research, including antiviral drug development.”
Blinking RNA
The work illuminates molecular-scale details of the virus’ activity inside host cells. In order to spread, viruses essentially take over cells and transform them into virus-producing factories, complete with special replication organelles. Within this factory, the viral RNA needs to duplicate itself over and over until enough genetic material is gathered up to move out and infect new cells and start the process over again.
The Stanford scientists sought to reveal this replication step in the sharpest detail to date. To do so, they first labeled the viral RNA and replication-associated proteins with fluorescent molecules of different colors. But imaging glowing RNA alone would result in fuzzy blobs in a conventional microscope. So they added a chemical that temporarily suppresses the fluorescence. The molecules would then blink back on at random times, and only a few lit up at a time. That made it easier to pinpoint the flashes, revealing the locations of the individual molecules.
Using a setup that included lasers, powerful microscopes, and a camera snapping photos every 10 milliseconds, the researchers gathered snapshots of the blinking molecules. When they combined sets of these images, they were able to create finely detailed photos showing the viral RNA and replication structures in the cells. “We have highly sensitive and specific methods and also high resolution,” said Leonid Andronov, co-lead author and Stanford chemistry postdoctoral scholar. “You can see one viral molecule inside the cell.”
The resulting images, with a resolution of 10 nanometers, reveal what might be the most detailed view yet of how the virus replicates itself inside of a cell. The images show magenta RNA forming clumps around the nucleus of the cell, which accumulate into a large repeating pattern. “We are the first to find that viral genomic RNA forms distinct globular structures at high resolution,” said Mengting Han, co-lead author and Stanford bioengineering postdoctoral scholar.
Video showing the different colored fluorescent labels blinking on and off, revealing more precise locations for individual molecules. | Leonid Andronov, Moerner Laboratory
The clusters help show how the virus evades the cell’s defenses, said W. E. Moerner , the paper’s co-senior author and Harry S. Mosher Professor of Chemistry in the School of Humanities and Sciences. “They’re collected together inside a membrane that sequesters them from the rest of the cell, so that they’re not attacked by the rest of the cell.”
Nanoscale drug testing
Compared to using an electron microscope, the new imaging technique can allow researchers to know with greater certainty where virus components are in a cell thanks to the blinking fluorescent labels. It also can provide nanoscale details of cell processes that are invisible in medical research conducted through biochemical assays. The conventional techniques “are completely different from these spatial recordings of where the objects actually are in the cell, down to this much higher resolution,” said Moerner. “We have an advantage based on the fluorescent labeling because we know where our light is coming from.”
Seeing exactly how the virus stages its infection holds promise for medicine. Observing how different viruses take over cells may help answer questions such as why some pathogens produce mild symptoms while others are life-threatening. The super-resolution microscopy can also benefit drug development. “This nanoscale structure of the replication organelles can provide some new therapeutic targets for us,” said Han. “We can use this method to screen different drugs and see its influence on the nanoscale structure.”
Indeed, that’s what the team plans to do. They will repeat the experiment and see how the viral structures shift in the presence of drugs like Paxlovid or remdesivir. If a candidate drug can suppress the viral replication step, that suggests the drug is effective at inhibiting the pathogen and making it easier for the host to fight the infection.
The researchers also plan to map all 29 proteins that make up SARS-CoV-2 and see what those proteins do across the span of an infection. “We hope that we will be prepared to really use these methods for the next challenge to quickly see what’s going on inside and better understand it,” said Qi.
For more information
Acknowledgements: Additional Stanford co-authors include postdoctoral scholar Yanyu Zhu, PhD student Ashwin Balaji, former PhD student Anish Roy, postdoctoral scholar Andrew Barentine, research specialist Puja Patel, and Jaishree Garhyan, director of the In Vitro Biosafety Level-3 Service Center . Moerner is also a member of Stanford Bio-X and the Wu Tsai Neurosciences Institute, and a faculty fellow of Sarafan ChEM-H . Qi is also a member of Bio-X, the Cardiovascular Institute , the Maternal & Child Health Research Institute (MCHRI), the Stanford Cancer Institute, and the Wu Tsai Neurosciences Institute, an institute scholar at Sarafan ChEM-H , and a Chan Zuckerberg Biohub – San Francisco Investigator.
This research was funded by the National Institute of General Medical Sciences of the National Institutes of Health. We also acknowledge use of the Stanford University Cell Sciences Imaging Core Facility.
Taylor Kubota, Stanford University: [email protected]
Four U.S. CRISPR Trials Editing Human DNA to Research New Treatments
Breaking down how the gene editing technology is being used, for the first time in the United States, to treat patients with severe medical conditions
Lila Thulin
Former Associate Editor, Special Projects
/https://tf-cmsv2-smithsonianmag-media.s3.amazonaws.com/filer/bc/78/bc780a37-4f25-40c7-ac3b-09de377ab51c/istock-952017558.jpg)
Last fall, the birth of genetically edited twin girls in China —the world’s first “designer babies”—prompted an immediate outcry in the medical science community. The change to the twins’ genomes, performed using the gene editing technology CRISPR, was intended to make the girls more resistant to H.I.V. But the edited genes may result in adverse side effects , and the International Commission on the Clinical Use of Human Germline Genome Editing is currently working on stricter and less ambiguous guidelines for editing the DNA of human embryos as a response to the rogue experiment.
Human genetic engineering has also witnessed more regulated advances. In the past 12 months, four clinical trials launched in the United States to use CRISPR to treat and potentially cure patients of serious medical conditions.
CRISPR-Cas9 is a technology derived from single-celled prokaryotic microorganisms and is composed of guide strands of RNA as well as the Cas9 enzyme, which does the "cutting." It allows scientists to make changes at highly specific locations in a cell’s genetic code by removing or replacing parts of the genome . Even tiny changes to individual genes can fundamentally alter the function of a cell. CRISPR has been used to edit all types of organisms, from humans to corn , but clinical trials represent a stride toward turning the technology into a drug or medical treatment.

The clinical trials in the U.S. are Phase 1 and 2 trials, small studies designed to demonstrate the safety and efficacy of a potential treatment. Essentially, these make-or-break trials take a drug from the laboratory to test on real patients. They’re “the first requirement for a product to end up on the market,” says Saar Gill, an assistant professor at the University of Pennsylvania’s medical school who works on genetically-edited immune cells.
While some of the diseases CRISPR therapies aim to tackle have other treatments available, part of gene editing’s allure lies in the possibility of a more effective or even permanent fix. The four U.S. clinical trials involving CRISPR have the potential to tackle cancers such as melanoma and lymphoma, sickle cell disease, and even blindness.
“As complicated and expensive as [genetic editing] is, you really are talking about the potential to cure a disease or essentially halt its progress or its adverse effect on the body forever,” Gill says.
Editing Patients’ T Cells to Fight Cancer
The first clinical trial in the U.S. to use CRISPR in a treatment began last September. Led by University of Pennsylvania professor of medicine Edward Stadtmauer, it consists of genetically modifying patients’ own T cells—a type of immune cell that circulates in the blood—to make them more efficient at fighting certain kinds of cancer cells. The 18 patients will have types of relapsed cancer, like multiple myeloma or melanoma, that tend to overproduce an antigen called NY-ESO-1.
Once the T cells have been extracted from the patients’ blood, scientists will make several edits using CRISPR as well as a genetic modification technique derived from viruses like H.I.V. An added gene will cause the modified T cells to target cells with NY-ESO-1 as if it were a microscopic signal flare.
Another edit will stop T cells from producing proteins that could distract the cells from targeting NY-ESO-1. And researchers will also aim to turbo-boost the T cells by eliminating a protein called PD-1 that can prevent the T cells from killing cancer cells.
Patients will undergo chemotherapy to deplete their natural reserve of T cells, and then they’ll receive an infusion of the edited cells to replace them. The specific chemotherapy isn’t likely to affect the patients' cancers, so that step of the trial won’t complicate the study’s assessment of the usefulness of T cell therapy.
According to a spokesperson for Penn Medicine, two patients—one with multiple myeloma and one with sarcoma—have already begun treatment. The trial is scheduled to conclude in 2033 , and it will assess both safety (whether the edited T cell treatment leads to any negative side effects) and also efficacy (measured by outcomes such as whether the cancer disappears, the length of remission, and overall patient survival).
Boosting Fetal Hemoglobin in Patients With Sickle Cell Disease
A trial helmed by Massachusetts-based Vertex Pharmaceuticals and CRISPR Therapeutics is the first CRISPR-based clinical trial in the U.S. for a condition with a clear, heritable genetic basis: sickle cell disease. The recessive condition is caused by a single base-pair change, meaning that both copies of a patient’s affected gene differ by just one genetic “letter” from a normally functioning gene. Victoria Gray, a 34-year-old woman from Mississippi who was recently profiled by NPR , was the first patient to receive CRISPR-edited stem cells as part of the trial.
The disease, which occurs most frequently in people of African descent, affects a protein called hemoglobin, which plays a critical role in helping red blood cells carry oxygen to different tissues in the body. Sickle cell causes hemoglobin proteins to clump into long fibers that warp disc-shaped red blood cells into sickle shapes. The irregularly shaped blood cells are short-lived and can’t flow smoothly through blood vessels, causing blockages, intense pain and anemia.

Like the University of Pennsylvania T cell study, the sickle cell trial involves editing a patient’s own cells ex-vivo, or outside of the body in a lab. Stem cells are collected from the bloodstream and edited with CRISPR so they will pump out high levels of fetal hemoglobin, a protein that typically dwindles to trace levels after infancy. Fetal hemoglobin (HbF) is encoded by an entirely different gene than beta-globin, the part of hemoglobin that can cause red blood cells to sickle. Adults with sickle cell whose bodies naturally make more HbF often experience less severe symptoms. Fetal hemoglobin can take one or both of sickle hemoglobin’s spots in the four-part hemoglobin molecule, substantially lowering a cell’s likelihood of adopting a sickle shape.
The trial, slated to conclude in May 2022 , will destroy participants’ unedited bone marrow cells with chemotherapy and then inject edited stem cells through a catheter in a onetime infusion. Doctors will look for the treatment to generate 20 percent or more HbF in the bloodstream for at least three months. Fetal hemoglobin normally constitutes only around 1 percent of adults’ hemoglobin supply, but previous studies have shown that proportions of fetal hemoglobin above 20 percent can keep enough cells from sickling to significantly reduce symptoms, including severe pain episodes.
If successful, the therapy would offer another option for a disease with few available treatments. The only current cure for sickle cell disease is a bone marrow transplant, but, according to the National Heart, Blood, and Lung Institute , such transplants work best in children and the likelihood of finding a marrow donor match is low. Just two FDA-approved drugs for sickle cell currently exist, aimed at ameliorating the worst of patients’ symptoms, and one of them, hydroxyurea , also works by increasing fetal hemoglobin.
Editing Donor T Cells to Fight Lymphoma
The same companies behind the sickle cell treatment have also begun a trial to use CRISPR-edited T cells to treat non-responsive or relapsed non-Hodgkin’s lymphoma. This cancer of the lymphatic system plays a major role in the body’s immune response. Unlike the University of Pennsylvania trial, the study involves editing T cells from donors. The cells will be edited using CRISPR to target CD-19, a protein that marks B cells, which become malignant in some types of non-Hodgkin’s lymphoma. The edits also remove two proteins to stop a patient’s immune system from rejecting the donated T cells and to prevent the edited T cells from attacking non-cancerous cells.
A 2019 poster from the researchers explains that a prototype treatment in mice with acute leukemia stalled tumor growth for about 60 days. Additionally, lab tests showed that modified human T cells were successfully able to target and kill CD-19-marked cancer cells. For the clinical trial, which will eventually include a maximum of 95 participants, researchers will track how patients tolerate different doses of the T cell treatment and how many patients see their cancers shrink or disappear entirely. After the treatment is complete, scientists will keep tabs on patients and their survival and recurrence rates over the course of five years.
Editing Photoreceptor Cells to Treat Inherited Blindness
At the end of July, Cambridge, Massachusetts-based Editas Medicine, working with Irish company Allergan, announced that they’d begun enrollment in a clinical trial for EDIT-101, a treatment for a type of inherited childhood blindness known as Leber Congenital Amaurosis (LCA). It will be the first instance of a CRISPR clinical trial that conducts cellular editing within a human body, or in vivo. The trial will include about 18 participants, including patients as young as age 3, with a particular subset of LCA caused by a single genetic mutation that impairs photoreceptors. These cells in the eye convert light into signals for the brain to process.
The treatment comes in the form of an injection into the space behind the retina . A type of virus known as an adenovirus will “infect” the photoreceptor cells with DNA instructions to produce Cas9, the CRISPR enzyme , to cut the photoreceptor genome in specified locations. The edits change the photoreceptors’ DNA to fix the blindness-causing mutation, spurring the cells to regrow previously faulty light-sensing components, which should improve the patients’ vision.
Medical researchers aim to affect 10 percent or more of the targeted photoreceptor cells, the threshold that other research suggests is required to make a leap in visual acuity. Medical staff will measure patients’ vision in various ways, including an obstacle course featuring barriers with different contrast levels, a color vision test, the pupil’s response to light, and the person’s own assessment of visual change.
The EDIT-101 treatment has been tested in non-human primates and also in tiny samples of a donated human retina. In the human retina, the desired edit was made about 17 percent of the time, and scientists detected no unintended “off-target” changes.
The method of injecting a virus subretinally to treat LCA has been successful before. Jean Bennett and Albert Maguire’s treatment Luxturna doesn’t involve CRISPR, but it does use a similar viral injection to deliver a working copy of a malfunctioning gene to pigment cells in the retina. The work was recognized by Smithsonian magazine’s 2018 Ingenuity Award for life sciences.
The Future of CRISPR in Medicine
Early clinical trials are not without risks. In 1999, an 18-year-old participant named Jesse Gelsinger died in a Phase 1 gene therapy trial—a tragedy that still lingers over the field. Gelsinger had inherited a metabolic disorder, and like other patients in the trial, received an injection straight to his liver of the ammonia-digesting gene his body lacked. Four days later, multiple organs failed , and Gelsinger was taken off life support. After his death, investigations uncovered a tangle of ethical lapses . Critics said inadequate information had been provided about the study’s risks and pointed out that a key administrator at the University of Pennsylvania center behind the study had a financial conflict of interest.
Mildred Cho, a bioethicist and professor at the Stanford School of Medicine, sits on NExTRAC , the panel that advises the National Institutes of Health (NIH) on emerging biotechnologies. She says she’s “concerned that the factors at play in Jesse Gelsinger’s death have not actually been eliminated.” Specifically, Cho is wary of the risks of clinical trials moving too quickly in an environment where patients, physician-scientists and pharmaceutical companies alike are anxious to alleviate devastating medical conditions. “I think there’s a lot of pressuring pushing these new technologies forward, and at the same time, there’s more reluctance to regulate,” she says.
In the U.S., the current scientific consensus is that CRISPR is worth the risk, particularly to treat serious diseases with few alternative options. Other gene therapies have been successful before, like the cancer treatments Kymriah and Yescarta . But unlike most other gene editing techniques, CRISPR is relatively easy to engineer and use, opening up the floodgates for possible applications. The potential of tools like CRISPR to cure currently unfixable diseases represents a “massive paradigm shift from taking a pill for the rest of your life,” Gill says.
CRISPR is no miracle cure, yet. Larger trials must follow this preliminary work before the FDA can approve any new treatment. James Wilson, the former director of the University of Pennsylvania center that ran the trial in which Jesse Gelsinger died, said in a recent interview : “It’s going to be a long road before we get to the point where editing would be deemed safe enough for diseases other than those that have really significant morbidity and mortality.”
But for conditions that often prove deadly or debilitating, a little genetic engineering, done properly, could go a long way.
Get the latest Science stories in your inbox.
Lila Thulin | | READ MORE
Lila Thulin is the former associate web editor, special projects, for Smithsonian magazine and covers a range of subjects from women's history to medicine.
- See us on facebook
- See us on twitter
- See us on youtube
- See us on linkedin
- See us on instagram

Stanford Medicine-led study clarifies how ‘junk DNA’ influences gene expression
Changes to short, repetitive sequences in the genome have been linked to diseases like autism and schizophrenia. New revelations about how such changes increase and decrease gene expression may provide insight into these and other disorders.
September 26, 2023 - By Jennifer Welsh

A study led by researchers at Stanford Medicine have unraveled some of the mystery of how non-coding DNA changes the level of gene expression. Picture Office
For decades, scientists have known that, despite its name, “junk DNA” in fact plays a critical role: While the coding genes provide blueprints for building proteins, which direct most of the body’s functions, some of the noncoding sections of the genome, including regions previously dismissed as “junk,” seem to turn up or down the expression of those genes.
But it’s been unclear how certain noncoding regions influence gene-expression levels — that is, the number of times a gene is copied into RNA and used to make proteins.
Now, a new study by Polly Fordyce , PhD, associate professor of bioengineering and of genetics, and her colleagues has unraveled some of the mystery. Their discovery may help researchers understand complex genetic conditions, including autism, schizophrenia, cancer and Crohn’s disease.
“We’ve known for a while that short tandem repeats, or STRs, aren’t junk because their presence or absence correlates with changes in gene expression,” Fordyce said. “But we haven’t known how they exert these effects.”
Authors of the study, published Sept. 22 in Science , believe it’s the first to offer a roadmap to understanding how STR changes can impact gene expression.
An evolving view of ‘junk DNA’
STRs make up about 5% of the human genome. “Starting in the 1980s, researchers noticed that changes to these repetitive sequences can affect gene expression,” said the study’s lead author, Connor Horton, who was a technician in Fordyce’s lab. “That’s the trail of breadcrumbs we’ve followed.”
For the study, the researchers looked at how STRs interact with proteins called transcription factors. Transcription factors attach to noncoding DNA, regulating the expression of protein-coding genes.

Polly Fordyce
“Researchers have spent a lot of time characterizing these transcription factors and figuring out which sequences — called motifs — they like to bind to the most,” Fordyce said. But current models don’t adequately explain where and when transcription factors bind to noncoding DNA to regulate gene expression. Sometimes, no transcription factor is attached to something that looks like a perfect motif. Other times, transcription factors bind to stretches of DNA that aren’t motifs.
“To solve the puzzle of why transcription factors go to some places in the genome and not to others, we needed to look beyond the highly preferred motifs,” Fordyce said. “In this study, we’re showing that the STR sequence around the motif can have a really big effect on transcription factor binding, providing clues as to what these repeated sequences might be doing.”
To better understand the role of short tandem repeats in gene expression, the researchers stripped the mechanisms down to their basics: transcription factors and naked DNA. They used specialized assays designed in the Fordyce lab to run thousands of tiny experiments side by side, saving time and money.
The experiments compared how tightly transcription factors attached to thousands of DNA sequences — those with a preferred motif, those without one, and those surrounded by random sequences or by a wide variety of STRs.
“In the experiment we asked, ‘How do these changes impact the strength of transcription factor binding?’” Fordyce said. “We saw a surprisingly large effect. Varying the STR sequence around a motif can have up to a 70-fold impact on the binding.”
To discover how the DNA and transcription factors interacted, Horton made hundreds of mutated transcription factors. He saw that changes to the transcription factor’s DNA binding domain affected whether it recognized the motif and the STRs. The researchers concluded that the transcription factors directly interact with the repetitive genetic code, attaching to it and the motif with the DNA binding domain.
Models to help understand polygenic diseases
The large number — over 6,000 — of experiments the team ran made it possible to develop a model of the rules governing transcription-factor binding. Their findings could even help researchers understand and model interactions between other transcription factors and noncoding regions of DNA that regulate gene expression.
“We set out to study short tandem repeats. But the models we developed apply broadly to the entire regulatory landscape,” said Horton, now a graduate student at the University of California, Berkeley. “It helps us better understand how transcription factors bind to regulatory DNA, even when short tandem repeats aren’t involved.”
Models of how noncoding regulatory regions impact transcription factor binding can help researchers understand the role of these sequences in polygenic diseases. “It’s been known for some time that short tandem repeats are associated with increased or decreased risk of certain diseases,” Horton said. “We hypothesize that changes in the short tandem repeats between individuals lead to different amounts of transcription factor binding, which leads to changes in gene expression, which might be linked to these diseases.”
Through the years, genome-wide association studies have linked changes in STRs to various diseases. “But it wasn’t clear what to do with that information,” Horton said. “Our models can suggest experiments to understand how those short tandem repeats affect progression or risk of the disease.”
This study was funded by the National Institutes of Health (grants R01-GM117106-0 and DP2-GM123641), the National Science Foundation, the ChEM-H Chemistry-Biology Interface pre-doctoral training program and the Swedish Research Council.
Researchers from Stowers Institute for Medical Research and Duke University School of Medicine contributed to this work.
- Jennifer Welsh Jennifer Welsh is a freelance writer
About Stanford Medicine
Stanford Medicine is an integrated academic health system comprising the Stanford School of Medicine and adult and pediatric health care delivery systems. Together, they harness the full potential of biomedicine through collaborative research, education and clinical care for patients. For more information, please visit med.stanford.edu .
Hope amid crisis
Psychiatry’s new frontiers


Depression, schizophrenia and bipolar disorder linked with ancient viral DNA in our genome – new research
Research Fellow, King's College London
Professor of Immunology in Medicine, Cornell University
Senior lecturer, King's College London
Disclosure statement
Rodrigo Duarte received funding from the National Institutes of Health, USA.
Douglas Nixon receives funding from the National Institutes of Health, USA.
Timothy Powell receives funding from the National Institute for Health and Care Research, the National Institutes of Health, and the Medical Research Council.
King's College London provides funding as a member of The Conversation UK.
View all partners
Around 8% of human DNA is made up of genetic sequences acquired from ancient viruses . These sequences, known as human endogenous retroviruses (or Hervs), date back hundreds of thousands to millions of years – with some even predating the emergence of Homo sapiens .
Our latest research suggests that some ancient viral DNA sequences in the human genome play a role in susceptibility to psychiatric disorders such as schizophrenia, bipolar disorder and major depressive disorder.
Hervs represent the remnants of these infections with ancient retroviruses. Retroviruses are viruses that insert a copy of their genetic material into the DNA of the cells they infect. Retroviruses probably infected us on multiple occasions during our evolutionary past. When these infections occurred in sperm or egg cells that generated offspring, the genetic material from these retroviruses was passed on to subsequent generations , becoming a permanent part of our lineage.
Initially, scientists considered Hervs to be “junk DNA” – parts of our genome with no discernible function. But as our understanding of the human genome has advanced, it’s become evident that this so-called junk DNA is responsible for more functions than originally hypothesised.
First, researchers found that Hervs can regulate the expression of other human genes. A genetic feature is said to be “expressed” if its DNA segment is used to produce RNA (ribonucleic acid) molecules. These RNA molecules can then serve as intermediaries leading to the production of specific proteins , or help to regulate other parts of the genome .
Initial research suggested that Hervs regulate the expression of neighbouring genes with important biological functions. One example of this is a Herv that regulates the expression of a gene involved in modifying connections between brain cells.
Hervs have also been found to produce RNAs and even proteins in blood and brain samples . These molecules have the potential to exert a wide range of functions, as they can travel across cellular compartments to execute different roles.
Scientists have also found evidence suggesting certain human genes are derived from Hervs. This indicates there were instances during evolution where Hervs were co-opted for specialised biological functions. For example, the human genes syncytins 1 and 2, which are derived from Hervs, play pivotal roles in placental development .
HERVs in psychiatric disorders
Considering the abundance of Hervs in the genome and their potentially numerous functions, we wanted to better understand whether genetic susceptibility to certain psychiatric disorders was associated with differences in Herv expression.

In our study , we profiled Herv expression in nearly 800 autopsy brain samples. This helped us identify DNA variations that influenced Herv expression in the brain.
We then cross-referenced this information with findings from large genetic studies which had compared genetic differences between tens of thousands of people – both with and without mental health conditions. These studies identified variations in DNA associated with different psychiatric conditions.
We found that that the expression of four Hervs was linked with genetic susceptibility to major psychiatric disorders. The expression of two of these Hervs was associated with schizophrenia, one Herv with both schizophrenia and bipolar disorder, and one with depression. These results suggest that Hervs may be playing a more important role in the brain than initially thought.
Read more: Discovering how genetic 'dark matter' plays a role in mental illness is just the tip of the iceberg for human health
There are many genes involved in psychiatric disorders – and Hervs are only a part of this puzzle. Although the precise impact of these Hervs on brain cells and on a person’s susceptibility to certain psychiatric disorders requires further research, our study is the first to show that genetic susceptibility for a psychiatric disorder also acts through these ancient viral DNA sequences.
It’s still too early to determine the practical applications of our findings – and whether they might be used to develop new treatments. But we’re optimistic about this line of research. By linking Herv expression in the brain with psychiatric disorders, our research recognises the importance of these mysterious sequences in the human genome, which have been ignored for years.
- Schizophrenia
- Bipolar disorder
- Retroviruses
- Psychiatric disorders

Head of School, School of Arts & Social Sciences, Monash University Malaysia

Chief Operating Officer (COO)

Clinical Teaching Fellow

Data Manager

Director, Social Policy

An official website of the United States government
Here’s how you know
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock A locked padlock ) or https:// means you’ve safely connected to the .gov website. Share sensitive information only on official, secure websites.
https://www.nist.gov/news-events/news/2023/01/new-dna-biosensor-could-unlock-powerful-low-cost-clinical-diagnostics
New DNA Biosensor Could Unlock Powerful, Low-Cost Clinical Diagnostics
DNA can signal the presence of or predisposition to a slew of diseases, including cancer. The ability to flag down these clues, known as biomarkers, allows medical professionals to make critical early diagnoses and provide personalized treatments. The typical methods of screening can be laborious, expensive or limited in what they can uncover. A new biosensor chip that boasts an accurate and inexpensive design may increase accessibility to high-quality diagnostics.
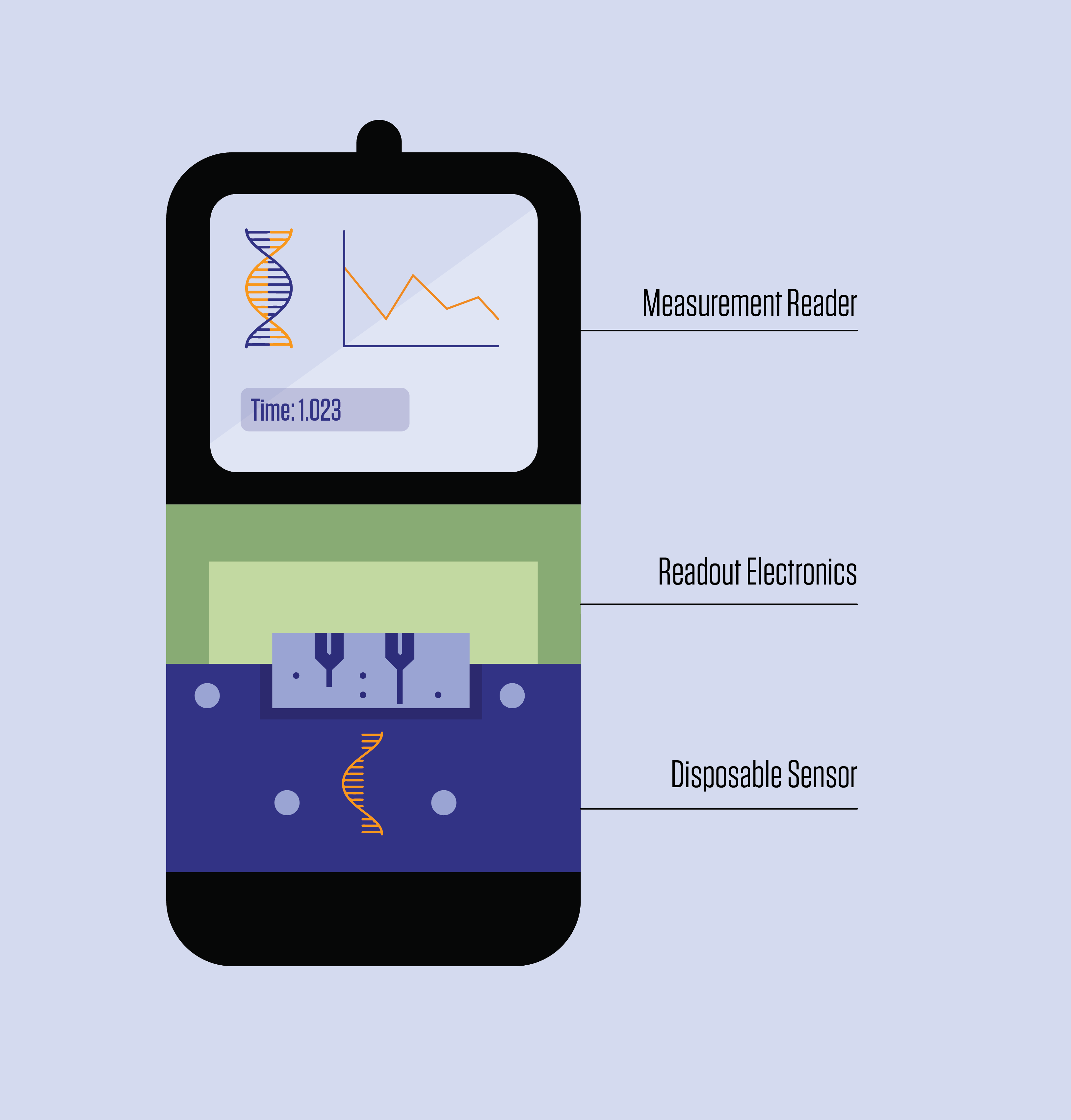
The biosensor, developed by researchers at the National Institute of Standards and Technology (NIST), Brown University and the French government-funded research institute CEA-Leti, identifies biomarkers by measuring how binding occurs between DNA strands and the device. What sets it apart from other similar sensors is its modular design, which lowers costs by making it easier to mass produce and allowing the most expensive components to be reused.
In a paper just posted online from the latest IEEE International Electron Devices Meeting, the researchers presented results of a study that demonstrates the device’s high sensitivity and precision despite its modularity, which is typically associated with diminished performance.
Like other DNA biosensors, the device takes advantage of the fact that a single DNA strand, when not paired with another within the familiar double helix, is primed for chemical bonding. Part of the device is coated with single strands of DNA. When these “probes” encounter DNA biomarkers that have a corresponding, or complementary, genetic sequence, the two strands bind, sending a signal that is picked up by the device.
“To make the measurement, we need two DNA molecules. We place one strand on our sensor that is complementary to the target DNA, that’s the proverbial needle in the haystack,” said NIST researcher Arvind Balijepalli, a co-author of the new study.
When a strand of target DNA binds to a probe, it induces a voltage shift that a semiconductor device, called a field-effect transistor (FET), can measure. These voltage shifts can occur hundreds of times a second as the molecules pop on and off the sensor.
Because of its high time resolution, this approach can tell you not only whether a DNA strand is bound to a probe, but how long it takes to connect and disconnect — a factor called binding kinetics that is key for discerning different markers that may bind to the same probe to varying degrees.
And with this method, you don’t need much space to measure a lot.
“This is a very scalable technique. In principle, we can have hundreds if not thousands of sensors in an area of one square millimeter integrated into a device the size of a smartphone, which is much less cumbersome than some of the technology currently used in the clinic,” Balijepalli said.
FET-based methods have yet to hit the mainstream, however. A significant stumbling block is their single-use nature, which until now has seemed a necessity but increases their cost.
Similar to how your radio becomes increasingly noisy as you drive away from a radio station, electrical signals also get noisier the longer they have to travel within electronics. The unwanted random noise picked up along the way makes the signal harder to measure.
To limit noise, DNA probes in FET-based sensors are normally attached to the transistor directly, which converts the signal into readable data. The drawback is that the probes are spent after being exposed to a sample, and thus the whole device is as well.
In the new study, Balijepalli and his colleagues increased the distance between the probes and the transistor so that the more expensive elements of the circuitry could be reused. The upfront penalty was that the distance could increase the amount of noise; however, there was much to be gained from the design choice, even beyond the cost savings.
“If the reader is reusable, we can build more sophisticated technology into it and get higher precision out of the readings, and it can interface with the inexpensive and disposable sensing element,” Balijepalli said.
Because they anticipated that the modular design would diminish the biosensor’s sensitivity, the researchers took a page out of the Internet of Things (IoT) playbook, which accommodates the losses associated with wireless devices. The NIST authors paired their circuitry with a specific type of extremely low-power FET developed at CEA-Leti that is used in smartwatches, personal assistants and other devices to amplify signals and compensate for the lost sensitivity.
To test the performance of their device, they placed it in liquid samples containing DNA strands associated with exposure to harmful ionizing radiation. Complementary DNA probes adorned electrodes wired to the FET. Across several samples, they varied the amount of target DNA.
The researchers found that the binding kinetics were sensitive enough to make accurate measurements even at low concentrations. Overall, the performance of the modular design matched that of integrated, nonmodular FET-based biosensors.
The next step in their research is to find out if their sensor can perform similarly with varying DNA sequences caused by mutations. Since many diseases are caused by or associated with mutated DNA, this capability is essential for clinical diagnostics.
Other studies may evaluate the sensor’s ability to detect genetic material associated with viruses, such as COVID-19, that could hint at infection.
In the meantime, the new technology could represent a viable foundation to build upon.
“There’s an opportunity to develop more sophisticated modular sensors that are much more accessible without sacrificing high quality measurements,” Balijepalli said.
Paper: Seulki Cho, Alexander Zaslavsky, Curt A. Richter, Jacob M. Majikes, J. Alexander Liddle, François Andrieu, Sylvain Barraud and Arvind Balijepalli. High-Resolution DNA Binding Kinetics Measurements with Double Gate FD-SOI Transistors. 2022 International Electron Devices Meeting. Published online Jan. 23, 2023. DOI: 10.1109/IEDM45625.2022.10019493
- Share full article
Advertisement
Supported by
New DNA Analysis Supports an Unrecognized Tribe’s Ancient Roots in California
A collaboration between researchers and the Muwekma Ohlone Tribe offers new evidence that their ancestors have lived in the Bay Area for thousands of years.

By Sabrina Imbler
In “The Handbook of the Indians of California ,” published in 1925, the anthropologist Alfred Kroeber pronounced the Ohlone people “extinct so far as all practical purposes are concerned,” noting that only “a few scattered individuals survive.”
Although the anthropologist would not recant his declaration of extinction until the 1950s, “the damage was done,” said Charlene Nijmeh, the chairwoman of the Muwekma Ohlone Tribe, which is far from extinct.
The Muwekma Ohlone Tribe is one descendant community of the Ohlone people, who originally lived on 4.3 million acres in the Bay Area. For decades, the Muwekma have sought to regain federally recognized status, which they lost in the 1920s. Linguistic scholars and archaeologists had suggested the Ohlone people migrated there 1,500 to 1,000 years ago. But the tribe has long asserted that its presence in the region goes back further.
A study published in March in the journal Proceedings of the National Academy of Sciences offers fresh genomic evidence that the Muwekma’s connection to the Bay Area goes back at least 2,000 years. Working alongside the Muwekma Ohlone Tribe, researchers from several universities extracted DNA from 12 ancient people buried in the region as far back as 2,000 years ago and found biological continuity with DNA collected from modern-day members of the tribe.
“Validation, finally,” said Monica Arellano, the vice chairwoman of the tribe and an author of the paper. “This adds to all the information we’ve put out there, years of compiling and doing research proving who we are.”
Alyssa Bader, an anthropologist at the University of Colorado Boulder who was not involved with the research, praised how the paper included representatives of the tribe as co-authors and foregrounded community engagement. “It’s recognized as a really essential part of the scientific research method and not something that’s buried in supplemental information,” Dr. Bader said, adding, “Those are all sort of exciting directions for genomic research that includes genomic research on Indigenous communities and ancestors.”
In 2014, the San Francisco Public Utilities Commission proposed a construction project at an archaeological site that was likely to have human burials. The commission reached out to the Muwekma Ohlone Tribe, the most probable descendants of the ancient people.
The tribe brought on the Far Western Anthropological Research Group, a cultural resources consulting firm, to conduct excavations at the site, which they named Síi Túupentak (Place of the Water Round House Site). Síi Túupentak, which sits near the confluence of the Alameda Creek and the Arroyo de la Laguna, was a lush site. The village community fished in the creeks and managed nearby forests and grasslands with controlled burning, said Brian Byrd, an archaeologist with Far Western.
Far Western also excavated another ancient site nearby, named Rummey Ta Kuččuwiš Tiprectak (Place of the Stream of the Lagoon Site), which was inhabited as early as 2,500 years ago.
Tribal members led the excavations of the burials, which was an emotionally challenging task. “It’s unfortunate that we had to move them,” Ms. Arellano said. “But if anybody has to do it, we take that responsibility very seriously and with as much care and love as possible.” The burials were most likely prepared for people of high lineage, as many were interred with precious shells, such as abalone pendants, Ms. Arellano said.
Occasionally when burials were discovered, the excavators gathered under a big tree and talked through the process to ensure everyone was heard, according to Dr. Byrd. “Trust has often been something that isn’t the first step these days for archaeology and native communities,” he said.
The Muwekma Ohlone tribal council wanted to know if the burials at the sites could help prove their people’s ancient presence in the Bay Area, said Alan Leventhal, the tribe’s archaeologist and an emeritus lecturer at San Jose State University.
Ripan Malhi, an anthropologist at the University of Illinois at Urbana-Champaign, and Noah Rosenberg, a population geneticist at Stanford University, joined the project to lead the ancient DNA analysis. The researchers presented all additional tests to the tribal council for approval. The council gave the researchers the go-ahead to study dental plaque for signs of inhalants such as tobacco, as well as to conduct tests to determine the sexes of the buried children, Dr. Byrd said. “We were able to identify a few samples from ancestors that had really good preservation,” Dr. Malhi said.
After extracting DNA at the two sites from 12 individuals who lived between several hundred and 1,900 years ago, the researchers compared their genomes with publicly available genetic information about other Indigenous communities in the Americas and ancient individuals worldwide. The oldest and most recent burials shared distinctive combinations of genetic variants, suggesting they were from related groups.
The analysis identified a shared component of ancestry linking people at the two ancient sites to modern-day members of the Muwekma. This ancestry can be found in other modern communities, but it is present in a much higher proportion in the Muwekma.
“It was surprising to find this level of continuity given the many disruptions the Ohlone people experienced during Spanish occupation, such as forced relocations and admixture with other tribes forcibly displaced by the Spanish,” Dr. Rosenberg said.
In keeping with the principles of Indigenous data sovereignty, the Muwekma will review requests for genomic data collected from the sites and tribal members, retaining power over how the data is used. “It’s minimizing potential harms to communities,” Dr. Bader said. “That is important.”
For members of the Muwekma Ohlone Tribe, these genetic results represent a new line of evidence that aligns with their tribe’s oral history. “That’s pretty much our story, having to prove who we are,” Ms. Arellano said. “We knew who we are, we know who we are, and we’re still here.”
The Muwekma can trace their ancestry through several missions in the Bay Area and resided on small settlements called rancherias until the early 1900s, Mr. Leventhal said.
The tribe had once been federally recognized under a different name, the Verona Band of Alameda County. But it lost recognition after 1927, when a superintendent from Sacramento determined that the Muwekma and more than 100 other tribal bands did not need land, effectively terminating the tribe’s formal federal recognition, Mr. Leventhal said. “The tribe was never terminated by any act of Congress,” he added.
The Muwekma hope the new study and any further research will strengthen their case for federal recognition. “The cost of living is pushing us out,” Ms. Nijmeh, the tribe’s chairwoman, said. “Recognition means that we will be able to have a land base and have a community village and have our people stay on our lands in their rightful place.”
Síi Túupentak will soon open an interpretive center featuring some of the artifacts from the excavation, informational signage about the history of the tribal language and a replica of an eagle, an allusion to the Muwekma creation story.
But the ancient people once buried at Síi Túupentak will be reinterred elsewhere, as close as possible to their original graves, Ms. Arellano said.
“That was supposed to be their final resting place,” she said. “They were never supposed to be moved.”
Sabrina Imbler is a reporter covering science and the environment. More about Sabrina Imbler
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
Genetics research articles from across Nature Portfolio
Genetics research is the scientific discipline concerned with the study of the role of genes in traits such as the development of disease. It has a key role in identifying potential targets for therapeutic intervention and also in understanding genetically based variations in response to therapeutic interventions.
Latest Research and Reviews
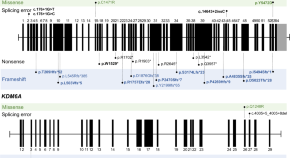
Clinical and molecular characteristics of Korean patients with Kabuki syndrome
- Ji-Hee Yoon
- Soojin Hwang
- Beom Hee Lee
Loss-of-function variants in ERF are associated with a Noonan syndrome-like phenotype with or without craniosynostosis
- Maria Lisa Dentici
- Marcello Niceta
- Marco Tartaglia
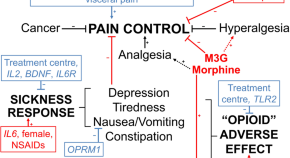
Pharmacokinetic and neuroimmune pharmacogenetic impacts on slow-release morphine cancer pain control and adverse effects
- Daniel T. Barratt
- Pål Klepstad
- Andrew A. Somogyi
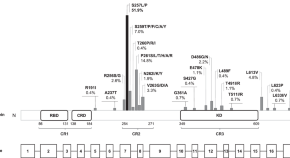
Defining the variant-phenotype correlation in patients affected by Noonan syndrome with the RAF1 :c.770C>T p.(Ser257Leu) variant
- Andrea Gazzin
- Federico Fornari
- Alessandro Mussa

Structural variant calling and clinical interpretation in 6224 unsolved rare disease exomes
- German Demidov
- Steven Laurie
- Stephan Ossowski
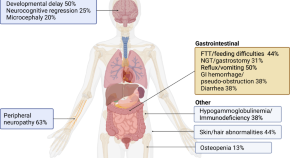
Genome sequencing enables diagnosis and treatment of SLC5A6 neuropathy
- Lisa G. Riley
- Subrata Sabui
- Shanti Balasubramaniam
News and Comment
The 1+Million Genomes Minimal Dataset for Cancer
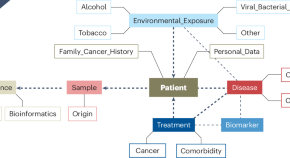
Defining minimal standards for data collection is key to creating interoperative, searchable genomic and clinical databases. We highlight here the 1+Million Genomes Minimal Dataset for Cancer, encompassing 140 items in 8 domains to foster the collection of cancer data, inform transnational cooperation and advance precision cancer medicine.
- Michela Riba
- Cinzia Sala
- Giovanni Tonon
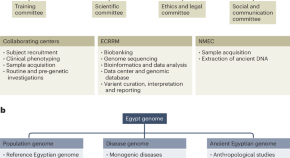
The Egypt Genome Project
The recently launched Egyptian Genome Project aims to sequence genomic variants of 100,000 apparently healthy Egyptian adults, with around 8,000 individuals suspected to have a genetic disease, as well as 200 ancient Egyptian mummies. The project will provide the first comprehensive genomic dataset from Egypt and North Africa.
- Mohamed A. Elmonem
- Neveen A. Soliman
- Khaled Amer
4D nucleome: dynamic three-dimensional genome organization over time
- Hyoung-Pyo Kim
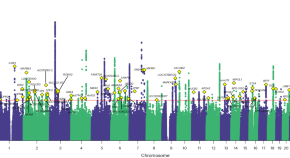
Deciphering the genetic architecture of pain intensity
Chronic pain is common, with more than one in five adult Americans reporting having pain daily or on most days. A multi-ancestry genomic analysis in 598,339 military veterans in the USA identifies 125 genetic variants associated with pain intensity, highlights shared genetic risk with substance use and psychiatric disorders, and reveals enrichment in GABAergic neurons as a key molecular contributor to experiencing pain.
Guidance on use of race, ethnicity, and geographic origin as proxies for genetic ancestry groups in biomedical publications
- W. Gregory Feero
- Robert D. Steiner
- Kirsten Bibbins-Domingo
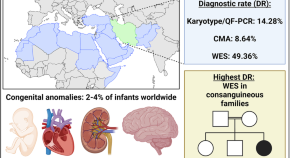
Optimizing genetic testing strategies for congenital anomalies in Iran
- Daniel G. Calame
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies

New Insights: Reversible Molecular Alterations Could Lead to Cancer, Research Reveals : ScienceAlert
C ancer affects half of the population at some point in their lives, and despite massive strides in understanding the disease, mysteries linger. Pioneering research is continually shedding light on cancer biology, including a groundbreaking discovery published recently which could revolutionize our conception of cancer genesis.
The longstanding belief has been that DNA mutations predominantly drive the transformation of healthy cells into cancerous ones. As we age, and under the influence of lifestyle and environmental factors like smoking and UV exposure, our cells accumulate random genetic alterations. While most of these alterations are inconsequential or result in cell death, some mutations aid cell survival. If a cell acquires enough of these “survival” mutations, it can start an unstoppable cycle of proliferation — the hallmark of cancer. This mutation-centric view of cancer is backed by ample evidence .
However, mutations are permanent changes that are typically challenging to tackle with medical interventions. If cancerous growths were solely due to genetic mutations, our treatment options could be constrained.
Contrastingly, other theories advocate that cancer could originate from other processes, opening up potential new avenues for prevention and therapy.
One such alternative hypothesis has been explored in a breakthrough Nature study . The research utilized fruit flies, which share a large portion of genes with humans, to probe the role of epigenetic changes—reversible markers that regulate gene activity—in cancer development.
While genetics involve direct alterations to DNA sequence, epigenetics refers to modifications that indirectly influence gene expression. Consider DNA as a book of life with each gene as a sentence. Genetic mutations are akin to permanent edits with a pen, whereas epigenetic tweaks resemble temporary pencil underlinings or bookmarks that can be easily added or removed.
These reversible epigenetic markers are crucial for development and can act as intermediaries between the environment and our genetic material. Though once thought to be too transient to cause cancer, research indicates that cancer cells exhibit numerous epigenetic changes that can be just as impactful as mutations in promoting cell survival.
The implication is that cancer may arise from a combination of genetic and epigenetic alterations. Yet, we lacked the evidence that epigenetic changes, without the presence of DNA mutations, could initiate cancer on their own—until this recent Nature publication .
For the first time, the study confirmed that temporary epigenetic modifications can instigate cancer even in the absence of genetic mutations.
Advancements in Cancer Treatment
This revelation isn’t just academically stimulating; it may very well alter cancer treatment paradigms if corroborated by further research. If epigenetic alterations can induce cancer, we could witness the emergence of epigenetic treatments to combat this lethal ailment. This concept has already spurred decades of research and drug development efforts.
Epigenetic therapies aim to correct cancer cells by altering epigenetic marks, potentially restoring normal cellular function and halting unchecked growth. Some of these therapies have already received approval in certain nations for specific types of blood cancers and sarcomas, with others being trialed for more prevalent cancers like those of the breast and prostate.
In the realm of cancer detection, the epigenetic model may enhance diagnostics. Aberrant epigenetic markers shed by cancer cells can be detected in patients’ blood. My collaborators and I have introduced a blood test capable of detecting such markers from minute blood samples. Integrating genetic and epigenetic testing could substantially improve cancer detection accuracy.
Furthermore, combining epigenetic drugs with traditional modalities like surgery or radiotherapy might provide a more comprehensive treatment approach. Our group also posits that epigenetic drugs and diagnostics may one day enable highly personalized therapies, although that prospect is still on the horizon.
While the epigenetic perspective enhances our comprehension of cancer progression, it doesn’t invalidate the classic mutation-based theory. Rather, it broadens our understanding of this complex disease.
The path forward is to validate the epigenetic cancer theory in additional models, like human cell lines, furthering the evolution of precision medicine.
Francesco Crea , Senior Lecturer in Cancer Genetics, The Open University
This commentary is a reproduction of an article from The Conversation published under a Creative Commons license. View the initial write-up .
FAQs about Reversible Molecular Changes and Cancer
Are epigenetic changes a cause of cancer.
Recent research indicates that epigenetic alterations, which were traditionally considered too transient, can indeed cause cancer even in the absence of genetic mutations. This understanding is pivotal in the study of how cancer can develop.
Can epigenetic marks be reversed?
Yes, unlike genetic mutations, epigenetic marks are reversible. These are modifications that do not change the DNA sequence but rather affect gene activity and how the cell reads the DNA.
What does this new understanding of epigenetics mean for cancer treatment?
It suggests the potential for epigenetic therapies. Such treatments could reset the epigenetic marks on cancer cells, returning them to a more normal state that ceases their uncontrolled proliferation.
Are there any epigenetic therapies currently available?
Some epigenetic therapies have been approved for treating certain blood cancers and sarcomas, and many others are undergoing clinical trials for various types of cancer.
How might epigenetic changes improve cancer detection?
Epigenetic markers released into the blood by cancer cells could be detected by blood tests, thus providing a non-invasive method of diagnosing cancer. Combining this with genetic testing could increase the accuracy of cancer detection.

- Search Menu
- Sign in through your institution
- Advance Articles
Editor's Choice
- Information for authors
- Submission Site
- Open Access Options
- Why publish with the journal
- About DNA Research
- About the Kazusa DNA Research Institute
- Editorial Board
- Advertising and Corporate Services
- Journals Career Network
- Self-Archiving Policy
- Dispatch Dates
- Journals on Oxford Academic
- Books on Oxford Academic

Editor-in-Chief
Satoshi Tabata
About the journal
DNA Research is an internationally peer-reviewed journal which aims at publishing papers of highest quality in broad aspects of DNA and genome-related research …
Latest Articles

High-Impact Research Collection
Explore a collection of freely available high-impact research from 2020 and 2021 published in DNA Research .
Browse the collection

DNA Research is the official journal of Kazusa DNA Research Institute, published by Oxford University Press and supported by funding from Chiba Prefecture, Japan.

Why publish in DNA Research?
Growing Impact Factor, fully open access journal, low open access charges, and more.

Volume 26, Issue 6: TASUKE+: a web-based platform for exploring GWAS results and large-scale resequencing data
Read the Executive Editor’s commentary
Resource Articles: Genomes Explored

Email alerts
Register to receive table of contents email alerts as soon as new issues of DNA Research are published online.

Recommend to your library
Fill out our simple online form to recommend DNA Research to your library.
Recommend now

Committee on Publication Ethics (COPE)
This journal is a member of and subscribes to the principles of the Committee on Publication Ethics (COPE)
publicationethics.org

PubMed Central
This journal enables compliance with the NIH Public Access Policy Read more

- Open access
Open access options for authors.

Accepting high quality papers on broad aspects of DNA and genome-related research.
Related Titles

- Author Guidelines
Affiliations
- Online ISSN 1756-1663
- Copyright © 2024 Kazusa DNA Research Institute
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.

- June 3, 2024 | Unlocking Sweet Secrets: How Taste Perception Influences Glucose Metabolism
- June 3, 2024 | 21 New Laser Materials Uncovered in Groundbreaking Global Study
- June 3, 2024 | Does Fruit Really Fuel Brain Growth? New Study Challenges Old Ideas
- June 3, 2024 | Nature’s Painkiller: Natural Molecules Found in Cannabis Rival Morphine in Groundbreaking Study
- June 3, 2024 | Bioluminescence and Biodiversity: Insights From La Parguera’s Marine Ecosystem
New DNA Research Unlocks Secrets of Native Rodents’ Rat Race to New Lands
By Australian National University September 2, 2022

A smoky mouse ( Pseudomys fumeus ) in the Grampians-Gariwerd National Park, Victoria, Australia. Credit: David Paul/Museums Victoria
New research has mapped the DNA from more than 150 species of native rodents from across Australia, New Guinea, and Melanesian islands. It paints a clearer picture of how they’re related and how they ended up spreading across the Pacific.
Lead author Dr. Emily Roycroft from The Australian National University (ANU) said native rodents are a fascinating but often under-appreciated evolutionary group.
“There are over 150 species in Australia and New Guinea that aren’t found anywhere else in the world, like the rakali – or ‘water rat’ – that’s often seen swimming around Canberra’s lakes,” said Dr. Roycroft.
“Until now, we’ve known very little about the evolution and origin of native rodents, especially species in New Guinea.”
The research team used a new approach to get DNA from museum specimens up to 180 years old, including many extinct and elusive species.

A wild Rakali (also known as the Australian Water Rat).
“One specimen of Guadalcanal rat from the Solomon Islands dates back to the 1880s, and the species hasn’t been seen since. It’s listed as critically endangered, and very possibly already extinct. We were curious to revisit these old specimens using modern technology,” said Dr. Roycroft.
The research reveals that mountain formation in New Guinea five million years ago was the trigger for the spread of native rodents across the region. The expansion of New Guinea opened up new environments for rodents to adapt to, including through increased connectivity with Australia, the Solomon Islands, and the Maluku Islands.
“We’ve known for some time that Australia’s native rodents originated in Asia and arrived in our region via water – possibly a single pregnant animal floating across on a piece of driftwood. Now we have an accurate timeline for this, and an explanation for why we see so many species today,” said Dr. Roycroft.
“Our study shows native rodents are exceptional at colonizing new areas. When they first arrived in Australia they adapted to a lot of new environments – including the arid desert.
Having extra information about native rodents’ history could prove vital to the future of these species, according to Dr. Roycroft.
“Native rodents have a deep intrinsic value in our ecosystems. They’re ecosystem engineers; they aerate soil via burrowing and foraging and they help to disperse seeds and fungal spores,” she said.
“They also play a role in food webs as an important source for native predators, and in turn, they feed on plants, fungi, and smaller animals themselves.
“But they also have the highest extinction rate of any Australian mammal group, due to extreme habitat loss and introduced predators. If we lose even one native species, it can throw off the balance in an ecosystem.
“Understanding how our native rodents evolved and adapted will help us to conserve those we have left.”
The study has been published in the journal Current Biology .
Reference: “New Guinea uplift opens ecological opportunity across a continent” by Emily Roycroft, Pierre-Henri Fabre, Anna J. MacDonald, Craig Moritz, Adnan Moussalli and Kevin C. Rowe, 2 September 2022, Current Biology . DOI: 10.1016/j.cub.2022.08.021
The research was supported by funding from Bioplatforms Australia through the Australian Government National Collaborative Research Infrastructure Strategy (NCRIS).
More on SciTechDaily

Discovery of Ancient Singing Dog Species May Teach Us About Human Vocalization

Forget Mammoths – These Scientists Are Working To Resurrect the Extinct Christmas Island Rat Through DNA Editing

“Giant Cloud Rat” Fossils Discovered in Philippine Caves

Shock Find Brings Extinct “Shark Bay” Mouse Back From the Dead

Electronically Linked Brains of Rats Communicate Directly to Solve Behavioral Puzzles

Rats Will Help Other Rats
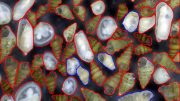
Native Biodiversity Collapse in the Eastern Mediterranean – Most Native Species Are Going Locally Extinct

Alien Species Invasions in Antarctica Predicted
Be the first to comment on "new dna research unlocks secrets of native rodents’ rat race to new lands", leave a comment cancel reply.
Email address is optional. If provided, your email will not be published or shared.
Save my name, email, and website in this browser for the next time I comment.

COMMENTS
DNA (deoxyribonucleic acid) is the nucleic acid polymer that forms the genetic code for a cell or virus. ... Research Highlights 17 May 2023 Nature Structural & Molecular Biology. ... A new study ...
In recent years, technologies that allow scientists to study a person's DNA at single-molecule resolution have vastly expanded our knowledge of the human genome, the microbiome, and the genetic ...
Within those new sequences of DNA, the scientists discovered more than 2,000 new genes. ... Environmental DNA research has aided conservation, but scientists say its ability to glean information ...
According to Garvan's Mahdi Zeraati, the first author of the new study, the i-motif is only one of a number of DNA structures that don't take the double helix form - including A-DNA, Z-DNA, triplex DNA and Cruciform DNA - and which could also exist in our cells. Another kind of DNA structure, called G-quadruplex (G4) DNA, was first ...
Published May 10, 2023 Updated May 12, 2023. More than 20 years after scientists first released a draft sequence of the human genome, the book of life has been given a long-overdue rewrite. A more ...
Now. The first gene-editing cure has arrived. Grateful patients are calling it "life changing.". It was only 11 years ago that scientists first developed the potent DNA-snipping technology ...
By University of California - Los Angeles Health Sciences August 12, 2023. Researchers at UCLA discovered a strong correlation between DNA methylation patterns and mammalian lifespan. Through extensive analysis, they found that DNA methylation, an epigenetic modification, plays a pivotal role in aging, with its effects visible across species ...
New technique will allow programmable manipulation of large DNA segments. A team of researchers led by Harvard and Broad Institute scientists has developed twin prime editing, a new, CRISPR-based gene-editing strategy that enables manipulation of gene-sized chunks of DNA in human cells without cutting the DNA double helix.
A new, nano-scale look at how the SARS-CoV-2 virus replicates in cells may offer greater precision in drug development, a Stanford University team reports in Nature Communications.Using advanced ...
The new research was published on May 17, in the journal Nature. Although it is widely understood that Homo sapiens originated in Africa, uncertainty surrounds how branches of human evolution diverged and how people migrated across the continent, said Brenna Henn, professor of anthropology and the Genome Center at UC Davis, corresponding author ...
DNA nanotechnology is a branch of nanotechnology concerned with the design, study and application of synthetic structures based on DNA. DNA nanotechnology takes advantage of the physical and ...
Contemporary DNA evidence suggests that humans emerged from the interaction of multiple populations living across the continent.. A new study in Nature challenges prevailing theories, suggesting that Homo sapiens evolved from multiple diverse populations across Africa, with the earliest detectable split occurring 120,000-135,000 years ago, after prolonged periods of genetic intermixing.
Four U.S. CRISPR Trials Editing Human DNA to Research New Treatments. Breaking down how the gene editing technology is being used, for the first time in the United States, to treat patients with ...
The researchers trained new neural networks that learn how the presence of an important DNA modification called 5hmC, either near the gene or far away from it, is related to gene expression activity.
In 1987, the New York Times Magazine characterized the Human Genome Project as the "biggest, costliest, most provocative biomedical research project in history." 2 But in the years between the ...
For decades, scientists have known that, despite its name, "junk DNA" in fact plays a critical role: While the coding genes provide blueprints for building proteins, which direct most of the body's functions, some of the noncoding sections of the genome, including regions previously dismissed as "junk," seem to turn up or down the expression of those genes.
For the first time ever, scientists have just discovered a new shape of DNA lurking inside human cells. In a study published Monday in Nature Chemistry, a team of researchers from the Kinghorn ...
Our latest research suggests that some ancient viral DNA sequences in the human genome play a role in susceptibility to psychiatric ... and whether they might be used to develop new treatments. ...
January 24, 2023. In a new study, researchers demonstrate the capability of DNA biosensor components for a unique modular DNA biosensor. The researchers plan to integrate their design within a device the size and shape of a smartphone for low-cost clinical diagnostics. DNA can signal the presence of or predisposition to a slew of diseases ...
After extracting DNA at the two sites from 12 individuals who lived between several hundred and 1,900 years ago, the researchers compared their genomes with publicly available genetic information ...
With our new technique, you place DNA molecules at defined locations on a surface and each protein from a mixture will then self-assemble to its corresponding DNA sequence, like an automated paint-by-number kit. ... CRISPR into a new realm of protein studies, and the molecular self-assembly strategy we show may assist in developing new research ...
RSS Feed. Genetics is the branch of science concerned with genes, heredity, and variation in living organisms. It seeks to understand the process of trait inheritance from parents to offspring ...
RSS Feed. Genetics research is the scientific discipline concerned with the study of the role of genes in traits such as the development of disease. It has a key role in identifying potential ...
Consider DNA as a book of life with each gene as a sentence. Genetic mutations are akin to permanent edits with a pen, whereas epigenetic tweaks resemble temporary pencil underlinings or bookmarks ...
The push from scientists is growing for a massive increase in research on RNA, a nucleic acid long in the shadow to the DNA that makes up genomes. Two days ago in The New York Times , Nobel laureate Thomas Cech published an opinion piece adapted from his new book titled The Catalyst: RNA and the Quest to Unlock Life's Deepest Secrets that ...
DNA Research is an internationally peer-reviewed journal which aims at publishing papers of highest quality in broad aspects of DNA and genome-related research ... Register to receive table of contents email alerts as soon as new issues of DNA Research are published online. Sign up . Recommend to your library.
A new type of blood test can predict the recurrence of breast cancer in high-risk patients, months or even years before they relapse, research has shown. A team from The Institute of Cancer Research, London, used an ultra-sensitive liquid biopsy to detect the presence of tiny amounts of cancer DNA left in the body following treatment for early ...
Science, Space and Technology News 2024. Hot Topics . June 1, 2024 | Chilling Forecast: Jellyfish Set To Dominate Arctic Waters by 2050; June 1, 2024 | Beyond Traditional Targets: New Protein Research Shakes Up Alzheimer's Treatments; June 1, 2024 | New Earth-Sized Planet Discovered Orbiting a Star That Will Live 100 Billion Years; June 1, 2024 | Record-Breaking Migration - Silky Shark ...
New research has mapped the DNA from more than 150 species of native rodents from across Australia, New Guinea, and Melanesian islands. It paints a clearer picture of how they're related and how they ended up spreading across the Pacific. Lead author Dr. Emily Roycroft from The Australian National University (ANU) said native rodents are a fascinating but often under-appreciated evolutionary ...