Subscribe or renew today
Every print subscription comes with full digital access

Science News

The largest known genome belongs to a tiny fern
Though 'Tmesipteris oblanceolata' is just 15 centimeters long, its genome dwarfs humans’ by more than 50 times.
Plant ‘time bombs’ highlight how sneaky invasive species can be
Flowers may be big antennas for bees’ electrical signals, more stories in plants.

Big monarch caterpillars don’t avoid toxic milkweed goo. They binge on it
Instead of nipping milkweed to drain the plants’ defensive sap, older monarch caterpillars may seek the toxic sap. Lab larvae guzzled it from a pipette.

How air pollution may make it harder for pollinators to find flowers
Certain air pollutants that build up at night can break down the same fragrance molecules that attract pollinators like hawk moths to primroses.

Giant tortoise migration in the Galápagos may be stymied by invasive trees
An invasion of Spanish cedar trees on Santa Cruz Island may block the seasonal migration routes of the island's giant tortoise population.

On hot summer days, this thistle is somehow cool to the touch
In hot Spanish summers, the thistle Carlina corymbosa is somehow able to cool itself substantially below air temperature.

Ancient trees’ gnarled, twisted shapes provide irreplaceable habitats
Traits that help trees live for hundreds of years also foster forest life, one reason why old growth forest conservation is crucial.

Here’s why blueberries are blue
Nanostructures in a blueberry’s waxy coating make it look blue, despite having dark red pigments — and no blue ones — in its skin, a new study reports.

This weird fern is the first known plant that turns its dead leaves into new roots
Cyathea rojasiana tree ferns seem to thrive in Panama’s Quebrada Chorro forest by turning dead leaves into roots that seek out nutrient-rich soil.

A rare 3-D tree fossil may be the earliest glimpse at a forest understory
The 350-million-year-old tree, which was wider than it was tall thanks to a mop-top crown of 3-meter-long leaves, would look at home in a Dr. Seuss book.

How an invasive ant changed a lion’s dinner menu
An invasive ant is killing off ants that defend trees from elephants. With less cover, it’s harder for lions to hunt zebras, so they hunt buffalo instead.
Subscribers, enter your e-mail address for full access to the Science News archives and digital editions.
Not a subscriber? Become one now .

Scientists identify 100 important questions facing plant science
- No Comments
An international panel of scientists have identified 100 of the most important questions facing plant science.
What are the key research priorities that will help tackle the global challenges of climate change, the biodiversity crises and feed a growing population in a sustainable way? Ten years after these priorities were first debated and summarised by a panel of scientists and published in New Phytologist , the panel reflects on the changes to plant science and the progress made to address these research areas, published in a Letter in New Phytologist .
To re-evaluate research priorities, a new panel was formed in 2022 to provide an international perspective on the important areas for plant science research. This project, led by Prof. Claire Grierson (University of Bristol, UK),is a novel piece of research that gathered over 600 questions about plant science, from botanically curious members of the public to scientific and industrial leaders around the world. A team of 20 plant scientists from 15 nations were assembled to identify 100 of the most important questions facing plant science.
These 100 questions are published in a Viewpoint in New Phytologist and highlight how climate change, biodiversity loss, and interdisciplinary and international collaborations are critical global priorities across diverse plant science research fields. The study demonstrates how critically important plant scientists believe the fight against climate change is, highlights global disparities in science funding and showcases a diverse range of important future research topics.
This inclusive study demonstrates how a global community of plant scientists, with a wide range of expertise, view the strategic priorities for plant research and offers insight into how different areas of research are important to different global regions. The study emphasises how an inclusive, international exercise can be used to identify diverse research questions. As panellist Dr Shyam Phartyal (Nalanda University, India) said, “One of the most significant steps of this study is maintaining a high level of diversity – not only in question gathering but also in the selection of panellists from the Global South.”
Another panellist, Dr Ida Wilson (Stellenbosch University, South Africa), said “My work is solution driven and needs to directly address the challenges that farmers in South Africa and Africa face. As part of the Africa panel, I am also very proud of the African inputs, and grateful for the opportunity to have our voices heard. I mentor young scientists too, and the excitement the publication has generated is tangible”.
Prof. Claire Grierson said, “These two papers form a unique and valuable resource for researchers and newcomers to plant science, including collaborators on interdisciplinary projects, students and early career researchers, and for policy development”.
Together, these two papers provide an excellent introduction to how plant science is developing and the significance, range and depth of research that needs to be addressed.
Read the papers : New Phytologist and New Phytologist
Article source : New Phytologist Foundation via Eurekalert
Image credit : Pexels / Pixabay

Bronce Sponsor

Are you an early-career researcher?

The Early-Career Researcher International Network (ECRi) , is a collection of activities addressed to help you. Check them now!
Previous Post Natural seed banks cannot protect biodiversity from the effects of climate change
Next post exploring the polysaccharide composition of plant cell walls in succulent aloes, related posts.

Genetic study of cauliflower reveals its evolutionary history

Tracing the evolution of ferns' surprisingly sweet defense strategy
Plants and the animals that eat them have evolved together in fascinating ways, creating a dynamic interplay of survival strategies. Many plants have developed physical and chemical defenses to fend off herbivores. A well-known strategy in flowering plants is to produce nectar to attract "ant bodyguards." Recent research explores the evolution of this same defense strategy in ferns.
Jacob Suissa, an assistant professor at the University of Tennessee Knoxville, led the study in collaboration with Boyce Thompson Institute's fern expert, Fay-Wei Li, and Cornell University's ant expert, Corrie Moreau.
The study, recently published in Nature Communications , revealed that ferns and flowering plants independently evolved nectaries, specialized structures that secrete sugary rewards to attract ant bodyguards, around the same time in the Cretaceous period. This finding is significant as it suggests similar evolutionary dynamics shaped the development of ant-plant mutualisms across these two divergent lineages, separated by more than 400 million years.
"Our research highlights a fascinating example of convergent evolution, where ferns and flowering plants independently developed similar strategies to defend themselves against predation by recruiting ant defenders with nectaries," said Suissa.
By integrating phylogenetic data and comparative analyses, the research team discovered that nectaries originated concurrently in ferns and angiosperms, but ferns experienced a significant lag in diversification compared to their flowering plant counterparts. The study also revealed that ferns likely recruited ant defenders secondarily, tapping into pre-existing ant-angiosperm relationships as they transitioned from the forest floor to the canopy.
"The evolutionary history of fern nectaries not only demonstrates the complex relationships between plants and insects -- relationships that have been previously underestimated -- but also underscores the ability of ferns to adapt to ecological challenges," explained Suissa.
The research offers new insights into the evolutionary dynamics that shape plant-animal interactions. By understanding how ferns and flowering plants independently developed similar defense mechanisms, scientists can better appreciate the underlying principles governing biodiversity and ecosystem function. This study opens new avenues for exploring the evolutionary history of other plant traits and their ecological impact, reinforcing the importance of mutualistic relationships in the natural world.
- Endangered Plants
- Evolutionary Biology
- Ecology Research
- Pests and Parasites
- Invasive Species
- Plant defense against being eaten
- Flowering plant
- Deadly nightshade and related plants
- Plant sexuality
Story Source:
Materials provided by Boyce Thompson Institute . Original written by Aaron Callahan. Note: Content may be edited for style and length.
Journal Reference :
- Jacob S. Suissa, Fay-Wei Li, Corrie S. Moreau. Convergent evolution of fern nectaries facilitated independent recruitment of ant-bodyguards from flowering plants . Nature Communications , 2024; 15 (1) DOI: 10.1038/s41467-024-48646-x
Cite This Page :
Explore More
- Resting Brain: Neurons Rehearse for Future
- Observing Single Molecules
- A Greener, More Effective Way to Kill Termites
- One Bright Spot Among Melting Glaciers
- Martian Meteorites Inform Red Planet's Structure
- Volcanic Events On Jupiter's Moon Io: High Res
- What Negative Adjectives Mean to Your Brain
- 'Living Bioelectronics' Can Sense and Heal Skin
- Extinct Saber-Toothed Cat On Texas Coast
- Some Black Holes Survive in Globular Clusters
Trending Topics
Strange & offbeat.
100+ Botany Research Topics [Updated 2024]

Botany, the scientific study of plants, holds the key to understanding the intricate and fascinating world of flora that surrounds us. As we delve into the realm of botany research, we uncover a vast array of botany research topics that not only contribute specifically to our scientific knowledge but also play an important role in addressing real-world challenges.
In this blog, we will embark on a journey through the rich landscape of botany research, exploring various captivating topics that researchers are delving into.
How to Select Botany Research Topics?
Table of Contents
Selecting an appropriate and engaging botany research topic is a crucial step in the research process. Whether you are a student working on a thesis, a scientist planning a research project, or someone passionate about exploring the wonders of plant biology, the right choice of topic can significantly impact the success and enjoyment of your research.
Here are some guidelines on how to select botany research topics:
- Identify Your Interests:
- Start by reflecting on your own personal interests within the field of botany. Consider the aspects of plant biology that fascinate you the most.
- Whether it’s plant physiology, taxonomy, ecology, genetics, or any other subfield, choosing a topic aligned with your interests can make the research process more enjoyable.
- Review Literature:
- Conduct a thorough review and it will be of existing literature in botany. Explore recent research articles, journals, and books to identify gaps in knowledge, emerging trends, and areas where further investigation is needed.
- This can help you find inspiration and identify potential research questions.
- Consider Relevance:
- Assess the relevance of your chosen topic to the current state of botany and its applications. Consider how your research could contribute to addressing real-world challenges, advancing scientific knowledge, or informing practical solutions.
- Relevant research topics often garner more attention and support.
- Evaluate Feasibility:
- Evaluate all possible feasibility of your chosen topic in terms of available resources, time constraints, and research capabilities.
- Consider the accessibility of study sites, the availability of equipment and materials, and the level of expertise required. A feasible research topic is one that aligns with your resources and constraints.
- Collaborate and Seek Guidance:
- Discuss your ideas with mentors, professors, or colleagues in the field.
- Collaborative discussions can provide valuable insights, help refine your research questions, and guide you toward topics that align with current research priorities.
- Consider working with a professional academic editor to review your work after you’ve finished writing it.
- Explore Emerging Technologies:
- Consider incorporating emerging technologies and methodologies in your research. This not only adds a contemporary dimension to your study but also opens up new possibilities for exploration.
- Technologies like CRISPR-Cas9, high-throughput sequencing, and remote sensing have revolutionized botany research.
- Think Interdisciplinary:
- Botany often intersects with various other disciplines, such as ecology, genetics, molecular biology, environmental science, and more.
- Consider interdisciplinary approaches to your research, as this can lead to innovative and comprehensive insights.
- Address Global Challenges:
- Botany research can play a crucial role in addressing global challenges like climate change, food security, and biodiversity loss.
- Choosing a topic that contributes to solving or mitigating these challenges adds societal relevance to your work.
- Explore Local Flora:
- If applicable, explore the flora of your local region. Investigating plant species native to your area can have practical implications for local conservation, biodiversity studies, and environmental management.
- Stay Inquisitive and Open-Minded:
- Keep an open mind and stay curious. Scientific research often involves unexpected discoveries, and being open to exploration can lead to novel and exciting findings.
- Be willing to adapt your research questions based on your findings and new insights.
100+ Botany Research Topics For All Students
Plant physiology.
- The Role of Plant Hormones in Growth and Development
- Mechanisms of Photosynthesis: A Comprehensive Study
- Impact of Environmental Stress on Plant Physiology
- Water Use Efficiency in Plants: Regulation and Adaptation
- Nutrient Uptake and Transport in Plants
- Signaling Pathways in Plant Defense Mechanisms
- Regulation of Flowering Time in Plants
- Physiological Responses of Plants to Climate Change
- Role of Mycorrhizal Associations in Plant Nutrition
- Stress Tolerance Mechanisms in Halophytic Plants
Plant Taxonomy
- Phylogenetic Analysis of a Plant Family: Case Study
- Integrating Molecular Systematics in Plant Taxonomy
- Plant DNA Barcoding for Species Identification
- Revision of a Plant Genus: Taxonomic Challenges
- Cryptic Species in Plant Taxonomy: Detection and Implications
- Floristic Diversity in a Specific Geographic Region
- Evolutionary Trends in Angiosperms
- Ethnobotanical Contributions to Plant Taxonomy
- Application of GIS in Plant Taxonomy
- Conservation Status Assessment of Endangered Plant Species
Plant Ecology
- Ecosystem Services Provided by Plants
- Dynamics of Plant-Animal Interactions in a Habitat
- Impact of Invasive Plant Species on Native Flora
- Plant Community Composition Along Environmental Gradients
- Ecological Consequences of Plant-Pollinator Decline
- Microbial Interactions in the Rhizosphere
- Plant Responses to Fire: Adaptation and Recovery
- Climate Change Effects on Plant Phenology
- Restoration Ecology: Reintroducing Native Plants
- Plant-Soil Feedbacks and Ecosystem Stability
Plant Pathology
- Molecular Mechanisms of Plant-Pathogen Interactions
- Emerging Plant Diseases: Causes and Consequences
- Integrated Disease Management in Agriculture
- Fungal Pathogens: Diversity and Control Strategies
- Plant Immunity and Defense Mechanisms
- Resistance Breeding Against Viral Pathogens
- Bacterial Diseases in Crop Plants: Diagnosis and Management
- Impact of Climate Change on Plant Pathogen Dynamics
- Biocontrol Agents for Plant Disease Management
- Genetic Basis of Host Susceptibility to Plant Pathogens
Ethnobotany
- Traditional Medicinal Plants: Documentation and Validation
- Cultural Significance of Plants in Indigenous Communities
- Ethnobotanical Survey of a Specific Region
- Sustainable Harvesting Practices of Medicinal Plants
- Traditional Plant Use in Rituals and Ceremonies
- Plant-Based Foods in Indigenous Diets
- Ethnopharmacological Studies on Antimicrobial Plants
- Conservation of Ethnobotanical Knowledge
- Ethnobotanical Contributions to Modern Medicine
- Indigenous Perspectives on Plant Conservation
Genetic and Molecular Biology
- CRISPR-Cas9 Applications in Plant Genome Editing
- Epigenetics in Plant Development and Stress Response
- Functional Genomics of Plant Responses to Abiotic Stress
- Genetic Diversity in Crop Plants and its Conservation
- Genetic Mapping and Marker-Assisted Selection in Plant Breeding
- Genome Sequencing of Non-Model Plant Species
- RNA Interference in Plant Gene Regulation
- Comparative Genomics of Plant Evolution
- Genetic Basis of Plant Adaptation to Extreme Environments
- Plant Epigenome Editing: Methods and Applications
Plant Anatomy and Morphology
- Comparative Anatomy of C3 and C4 Plants
- Xylem and Phloem Development in Plants
- Leaf Anatomy and Adaptations to Photosynthesis
- Morphological Diversity in Plant Reproductive Structures
- Evolution of Floral Symmetry in Angiosperms
- Root Architecture and its Functional Significance
- Stem Cell Dynamics in Plant Meristems
- Comparative Morphology of Succulent Plants
- Tissue Regeneration in Plants: Mechanisms and Applications
- Wood Anatomy and Tree-Ring Analysis in Dendrochronology
Climate Change and Plant Responses
- Impact of Global Warming on Alpine Plant Communities
- Plant Responses to Elevated CO2 Levels
- Drought Tolerance Mechanisms in Plants
- Shifts in Plant Phenology Due to Climate Change
- Climate-Induced Changes in Plant-Pollinator Interactions
- Carbon Sequestration Potential of Forest Ecosystems
- Ocean Acidification Effects on Seagrass Physiology
- Plant Responses to Increased Frequency of Extreme Events
- Alpine Plant Adaptations to Harsh Environments
- Climate-Driven Changes in Plant Distribution and Biogeography
Emerging Technologies in Botany Research
- Application of Machine Learning in Plant Phenotyping
- Nanotechnology in Plant Science: Current Status and Future Prospects
- Metagenomics in Studying Plant Microbiomes
- Remote Sensing for Monitoring Plant Health
- High-Throughput Sequencing in Plant Genomics
- CRISPR-Based Gene Drives for Ecological Restoration
- Advances in Plant Imaging Techniques
- Synthetic Biology Approaches in Plant Engineering
- Augmented Reality Applications in Plant Biology Education
- Digital Herbariums: Integrating Technology in Plant Taxonomy
Misc Botany Research Topics
- Metabolic Pathways in Plant Secondary Metabolism: Regulation and Significance
- Population Genomics of Endangered Plant Species: Implications for Conservation
- Impact of Soil Microbes on Plant Health and Productivity
- Evolutionary Dynamics of Plant-Pathogen Coevolution: Insights from Molecular Data
- Application of CRISPR-Based Gene Editing for Improving Crop Traits
- Phytochemical Profiling of Medicinal Plants for Drug Discovery
- Investigating the Role of Epigenetic Modifications in Plant Stress Responses
- Role of Plant Volatile Organic Compounds (VOCs) in Ecological Interactions
- Biotic and Abiotic Factors Influencing Plant Microbiome Composition
- Molecular Basis of Plant-Microbe Symbiosis: Lessons from Nitrogen-Fixing Associations
How to Make Botany Research Successful?
Conducting successful botany research involves a combination of careful planning, effective execution, and thoughtful analysis. Whether you are a student, a researcher, or someone conducting independent studies, here are key tips to ensure the success of your botany research:
- Establish Clear Objectives: Clearly articulate the goals and objectives of your research. What specific inquiries do you intend to address? A well-defined research focus serves as a guiding framework, ensuring your efforts remain purposeful and on course.
- Conduct an In-Depth Literature Review: Immerse yourself in the existing body of literature within your field of study. Identify gaps, discern trends, and pinpoint areas where your research could contribute significantly. A thorough literature review lays a robust groundwork for shaping your research design.
- Choose an Appropriate Research Topic: Select a research topic that resonates with your interests, aligns with your expertise, and addresses the current needs of the scientific community. Ensure that the chosen topic is not only feasible but also harbors the potential for impactful outcomes.
- Develop a Sound Research Plan: Create a detailed research plan outlining the methodologies, timelines, and resources required. A well-structured plan helps in efficient execution and minimizes the risk of unforeseen challenges.
- Utilize Cutting-Edge Technologies: Stay updated with the latest technologies and methodologies in botany research. Incorporate advanced tools such as high-throughput sequencing, CRISPR-Cas9 , and remote sensing to enhance the precision and efficiency of your research.
- Collaborate and Seek Guidance: Collaborate with experts in the field, seek mentorship, and engage in discussions with colleagues. Networking and collaboration can provide valuable insights, guidance, and potential avenues for collaboration.
- Ensure Ethical Considerations: Adhere to ethical guidelines and standards in your research. Obtain necessary approvals for human subjects, follow ethical practices in plant experimentation, and ensure the responsible use of emerging technologies.
- Implement Robust Experimental Design: Design experiments with attention to detail, ensuring that they are replicable and provide statistically significant results. Address potential confounding variables and incorporate controls to enhance the reliability of your findings.
- Collect and Analyze Data Thoughtfully: Implement systematic data collection methods. Use appropriate statistical analyses to interpret your results and draw meaningful conclusions. Transparent and well-documented data analysis enhances the credibility of your research.
- Regularly Review and Adapt: Periodically review your progress and be open to adapting your research plan based on emerging findings. Flexibility and responsiveness to unexpected results contribute to a dynamic and successful research process.
- Communicate Your Research Effectively: Share your findings through publications, presentations, and other relevant channels. Effective communication of your research results contributes to the broader scientific community and enhances the impact of your work.
- Foster a Collaborative Research Environment: Encourage collaboration within your research team. A collaborative environment fosters creativity, diverse perspectives, and a collective effort towards achieving research goals.
- Contribute to Sustainable Practices: If your research involves fieldwork or plant collection, adhere to sustainable practices. Consider the impact on local ecosystems and strive to minimize any negative consequences.
- Stay Resilient: Research can have its challenges, setbacks, and unforeseen obstacles. Stay resilient, remain focused on your goals, and view challenges as opportunities for growth and learning.
- Celebrate Achievements and Learn from Failures: Acknowledge and celebrate your achievements, no matter how small. Learn from any setbacks or failures and use them as lessons to refine and improve your research approach.
In the vast and diverse field of botany research, scientists are continually unraveling the mysteries of the plant kingdom. From the intricate processes of photosynthesis to the challenges posed by emerging plant diseases and the potential of cutting-edge technologies, botany research is a dynamic and ever-evolving field.
As we delve deeper into the green secrets of the plant world, our understanding grows, offering not only scientific insights but also solutions to address pressing global challenges such as food security, biodiversity loss, and climate change.
The exploration of botany research topics is a journey of discovery, paving the way for a sustainable and harmonious coexistence with the plant life that sustains our planet.
Related Posts

Step by Step Guide on The Best Way to Finance Car

The Best Way on How to Get Fund For Business to Grow it Efficiently
- Search Menu
- Sign in through your institution
- Advance Articles
- Collections
- Focus Collections
- Teaching Tools in Plant Biology
- Browse by cover
- High-Impact Research
- Author Guidelines
- Quick and Simple Author Support
- Focus Issues Call for Papers
- Submission Site
- Open Access Options
- Self-Archiving Policy
- Why Publish with Us?
- About The Plant Cell
- About The American Society of Plant Biologists
- Editorial Board
- Advertising & Corporate Services
- Journals on Oxford Academic
- Books on Oxford Academic

Browse issues

Cover image

Volume 36, Issue 6, June 2024
Thank you to reviewers and editors for 2023.
- View article
From the archives: Tales from evolution—inflorescence diversity, gene duplication, and chromatin-mediated gene regulation
It takes two to tango: plant hosts influence bacterial effector function through post-translational modifications, find your way: building an atlas from transcription factor activities in the marchantia gemma, mining for phosphates: the myb73-1 - gdpd2 - ga2ox1 module as a hub to improve phosphate deficiency tolerance in soybean, the mrna puzzle: intron retention under stress, more pi, anyone interplay between brassinosteroid signaling and the phosphate starvation response, dicing dicer-like2 roles: identification of sirna-independent dicer-like2 functions, a compact topic: how ethylene controls crown root development in compacted soil, what is going on inside of phytochrome b photobodies.

CONSTANS, a HUB for all seasons: How photoperiod pervades plant physiology regulatory circuits

Breakthrough Report
Heat-induced sumoylation differentially affects bacterial effectors in plant cells.

Multiple effectors from the pathogen P. syringae are substrates of SUMOylation, which regulates distinct effectors differently in plant cells during bacterial infection under high temperatures.
- Supplementary data
Large-scale Biology
Pan-transcriptomic analysis reveals alternative splicing control of cold tolerance in rice.

Pan-transcriptomic analysis of rice under cold stress reveals alternative splicing control of cold tolerance.
The landscape of transcription factor promoter activity during vegetative development in Marchantia

A collection of promoters was systematically derived from transcription factor genes in Marchantia polymorpha and used to build a precise map of cell fates in vegetative propagules of the plant.
Integrative omics analysis elucidates the genetic basis underlying seed weight and oil content in soybean

Integration of multiple association studies reveals genetic loci, coexpression modules, and candidate genes that are significantly associated with simultaneous regulation of seed weight and oil content.
Research Articles
The myb73–gdpd2–ga2ox1 transcriptional regulatory module confers phosphate deficiency tolerance in soybean.

The major quantitative trait locus gene GmGDPD2 controls root and phosphorus efficiency traits and interacts with Myb73 and GA2ox1 to enhance yield traits.
Maternal nitric oxide homeostasis impacts female gametophyte development under optimal and stress conditions

Maternal nitric oxide homeostasis restricts auxin supply to the female gametophyte, disrupting its development under both optimal and stress conditions.
Arginine methylation of SM-LIKE PROTEIN 4 antagonistically affects alternative splicing during Arabidopsis stress responses

Arginine methylation of the spliceosome component SM-LIKE PROTEIN 4 by PROTEIN ARGININE METHYLTRANSFERASE5 fine-tunes alternative splicing of genes to control stress responses in Arabidopsis.
The chromatin remodeler ERCC6 and the histone chaperone NAP1 are involved in apurinic/apyrimidinic endonuclease-mediated DNA repair

APURINIC/APYRIMIDINIC ENDONUCLEASE REDOX PROTEIN recruits a chromatin remodeler and a histone chaperone to facilitate uracil N-glycosylase-triggered base excision repair.
Brassinosteroid-dependent phosphorylation of PHOSPHATE STARVATION RESPONSE2 reduces its DNA-binding ability in rice

Brassinosteroids modulate phosphate homeostasis by phosphorylating a serine residue of OsPHR2 through GSK2, thereby attenuating its DNA-binding activity.
Identification and functional characterization of conserved cis -regulatory elements responsible for early fruit development in cucurbit crops

Integrative analyses of conserved noncoding sequences and accessible chromatin regions provide insights into cis -regulatory elements and target sequences for fruit development in cucurbits.
Evidence for an RNAi-independent role of Arabidopsis DICER-LIKE2 in growth inhibition and basal antiviral resistance

Arabidopsis DICER-LIKE2 mediates growth repression and basal antiviral resistance in local infection contexts independently of the silencing activity of the small interfering RNAs it produces.
Nonspecific phospholipases C3 and C4 interact with PIN-FORMED2 to regulate growth and tropic responses in Arabidopsis

Nonspecific phospholipases NPC3 and NPC4 interact with the transporter PIN-FORMED2 and regulate auxin transport and signaling.
OPEN STOMATA 1 phosphorylates CYCLIC NUCLEOTIDE-GATED CHANNELs to trigger Ca 2+ signaling for abscisic acid-induced stomatal closure in Arabidopsis

Abscisic acid activates CYCLIC NUCLEOTIDE-GATED CHANNELs via OST1 kinase-mediated phosphorylation to elicit and regulate cytosolic Ca 2+ signaling for stomatal closure in Arabidopsis.
ENHANCER OF SHOOT REGENERATION1 promotes de novo root organogenesis after wounding in Arabidopsis leaf explants

The dual mode of action of ENHANCER OF SHOOT REGENERATION1, which occurs near wound sites of leaf explants, maximizes local auxin biosynthesis and de novo root organogenesis in Arabidopsis.
A gradient of the HD-Zip regulator Woolly regulates multicellular trichome morphogenesis in tomato

A gradient-dependent mechanism mediated by the HD-Zip IV regulator Woolly controls cell division and endoreduplication during tomato ( Solanum lycopersicum ) multicellular trichome morphogenesis.
The OsEIL1–OsWOX11 transcription factor module controls rice crown root development in response to soil compaction

The phytohormone ethylene fine-tunes rice crown root development by activating WUSCHEL-RELATED HOMEOBOX 11 expression in response to soil compaction.
H3K4me1 recruits DNA repair proteins in plants

DNA repair proteins preferentially repair regions with the epigenomic feature H3K4me1, resulting in mutation rate heterogeneity in plants.
Transcription factor FveMYB117a inhibits axillary bud outgrowth by regulating cytokinin homeostasis in woodland strawberry

An MYB transcription factor directly regulates cytokinin homeostasis to repress axillary bud growth and consequently affect crown formation in woodland strawberry.
Corrections
Correction to: h 2 o 2 -dependent oxidation of the transcription factor gmntl1 promotes salt tolerance in soybean, correction to: polyploid plants take cytonuclear perturbations in stride, email alerts.
- Recommend to Your Librarian
- Advertising & Corporate Services
- Awards & Funding
- Plant Science Today
- Plant Biology Meeting
- Meeting Management Services
- Plant Science Research Weekly
- Taproot: A Plantae Podcast
Affiliations
- Online ISSN 1532-298X
- Print ISSN 1040-4651
- Copyright © 2024 American Society of Plant Biologists
- About Oxford Academic
- Publish journals with us
- University press partners
- What we publish
- New features
- Open access
- Institutional account management
- Rights and permissions
- Get help with access
- Accessibility
- Advertising
- Media enquiries
- Oxford University Press
- Oxford Languages
- University of Oxford
Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide
- Copyright © 2024 Oxford University Press
- Cookie settings
- Cookie policy
- Privacy policy
- Legal notice
This Feature Is Available To Subscribers Only
Sign In or Create an Account
This PDF is available to Subscribers Only
For full access to this pdf, sign in to an existing account, or purchase an annual subscription.
23 Ideas for Science Experiments Using Plants
ThoughtCo / Hilary Allison
- Cell Biology
- Weather & Climate
- B.A., Biology, Emory University
- A.S., Nursing, Chattahoochee Technical College
Plants are tremendously crucial to life on earth. They are the foundation of food chains in almost every ecosystem. Plants also play a significant role in the environment by influencing climate and producing life-giving oxygen. Plant project studies allow us to learn about plant biology and potential usage for plants in other fields such as medicine, agriculture, and biotechnology. The following plant project ideas provide suggestions for topics that can be explored through experimentation.
Plant Project Ideas
- Do magnetic fields affect plant growth?
- Do different colors of light affect the direction of plant growth?
- Do sounds (music, noise, etc.) affect plant growth?
- Do different colors of light affect the rate of photosynthesis ?
- What are the effects of acid rain on plant growth?
- Do household detergents affect plant growth?
- Can plants conduct electricity?
- Does cigarette smoke affect plant growth?
- Does soil temperature affect root growth?
- Does caffeine affect plant growth?
- Does water salinity affect plant growth?
- Does artificial gravity affect seed germination?
- Does freezing affect seed germination?
- Does burned soil affect seed germination?
- Does seed size affect plant height?
- Does fruit size affect the number of seeds in the fruit?
- Do vitamins or fertilizers promote plant growth?
- Do fertilizers extend plant life during a drought?
- Does leaf size affect plant transpiration rates?
- Can plant spices inhibit bacterial growth ?
- Do different types of artificial light affect plant growth?
- Does soil pH affect plant growth?
- Do carnivorous plants prefer certain insects?
- 8th Grade Science Fair Project Ideas
- Plant and Soil Chemistry Science Projects
- High School Science Fair Projects
- Middle School Science Fair Project Ideas
- Animal Studies and School Project Ideas
- Environmental Science Fair Projects
- Elementary School Science Fair Projects
- College Science Fair Projects
- Chemistry Science Fair Project Ideas
- Magnetism Science Fair Projects
- 11th Grade Science Fair Projects
- 9th Grade Science Fair Projects
- Science Fair Project Ideas
- 4th Grade Science Fair Projects
- Caffeine Science Fair Projects
- Science Fair Experiment Ideas: Food and Cooking Chemistry
share this!
May 29, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
Researchers cataloging plant species are trying to decipher what makes some groups so successful
by Trinity College Dublin

Irish researchers involved in cataloging the world's plant species are hunting for answers as to what makes some groups so successful.
One of their major goals is to predict more accurately which lineages of flowering plants—some of which are of huge importance to people and to ecosystems—are at a greater risk from global climate change .
There are about 350,000 species of flowering plants on Earth, and each one is organized into a group called a genus, made up of closely related species with structural similarities. Some genera are small with just a single species (with relatively unique traits), but others are disproportionately large and contain thousands of species.
The "big plant genera" form a significant proportion of both global and Irish plant diversity, and are of disproportionate importance for both human nutrition and planetary health. Roughly 1 in 4 flowering plant species is a member of one of these.
Twenty years after the first assessment of big plant genera, research led by Trinity College Dublin and published today (May 29) in the Proceedings of the Royal Society B: Biological Sciences shows that these big genera are getting bigger and bigger as more and more species are described.
In fact, more than 10,000 species have been described in just 83 big genera since the year 2000, which is about 2.5 times the size of the total flora of Ireland.
Dr. Peter Moonlight, Assistant Professor in Trinity's School of Natural Sciences, led the just-published study. He said, "Until recently, big plant genera were seen as too large to study. But a recent revolution in methods in plant science and the development of global, collaborative networks has allowed us to update our understanding of plant evolution and global plant diversity.
"We now hope to identify common patterns across big plant genera that may explain why they are big when the other 99% of genera are small. Perhaps they have similar distributions, genetics, or morphology—we don't know yet, but this study is a key step to starting to understand this important evolutionary question.
"Big genera represent lineages of flowering plants that have been extremely successful in the game of evolution. Understanding why they became so successful may help us predict how they and other lineages on the tree of life will respond to the ongoing climate and biodiversity crises.
"Given that species in these big genera often have narrow ecological ranges in which they flourish, they may be more likely to be threatened by extinction as and when conditions change. They are a significant proportion of our global biodiversity, so perhaps we need to focus our conservation efforts most keenly on them."
Journal information: Proceedings of the Royal Society B
Provided by Trinity College Dublin
Explore further
Feedback to editors

Saturday Citations: The sound of music, sneaky birds, better training for LLMs. Plus: Diversity improves research
8 hours ago

Study investigates a massive 'spider' pulsar
10 hours ago

Greener, more effective termite control: Natural compound attracts wood eaters
12 hours ago

Shear genius: Researchers find way to scale up wonder material, which could do wonders for the Earth

New vestiges of the first life on Earth discovered in Saudi Arabia
May 31, 2024

Mussels downstream of wastewater treatment plant contain radium, study reports

A new way to see viruses in action: Super-resolution microscopy provides a nano-scale look

Martian meteorites deliver a trove of information on red planet's structure

New imager acquires amplitude and phase information without digital processing

AI helps scientists understand cosmic explosions
Relevant physicsforums posts, a dna animation, probability, genetic disorder related.
May 28, 2024
Looking For Today's DNA Knowledge
May 27, 2024
Covid Vaccines Reducing Infections
Human sperm, egg cells mass-generated using ips, and now, here comes covid-19 version ba.2, ba.4, ba.5,....
May 25, 2024
More from Biology and Medical
Related Stories
Global scientific network highlights plant genera named for women
Jan 8, 2024
One of the oldest land plant lineages, clubmosses (Selaginella), re-classified
Sep 13, 2023
Scientists present the first set of global maps showing geographic patterns of beta-diversity in flowering plants
Oct 16, 2023

State of the world's orchids revealed in new report
Oct 10, 2023

International study produces a comprehensive 'tree of life' for flowering plants
Apr 24, 2024

'Winners and losers' as global warming forces plants uphill
Mar 25, 2024
Recommended for you

This tiny fern has the largest genome of any organism on Earth

Clues to mysterious disappearance of North America's large mammals 50,000 years ago found within ancient bone collagen

Jumping spider study finds offspring care extends lifespan of mothers

The missing puzzle piece: A striking new snake species from the Arabian Peninsula
May 30, 2024

Trout in mine-polluted rivers are genetically 'isolated,' new study shows


Study shows cuckoos evolve to look like their hosts—and form new species in the process
Let us know if there is a problem with our content.
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Phys.org in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
Loading metrics
Open Access
Peer-reviewed
Meta-Research Article
Meta-Research Articles feature data-driven examinations of the methods, reporting, verification, and evaluation of scientific research.
See Journal Information »
Assessing the evolution of research topics in a biological field using plant science as an example
Roles Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing
* E-mail: [email protected]
Affiliations Department of Plant Biology, Michigan State University, East Lansing, Michigan, United States of America, Department of Computational Mathematics, Science, and Engineering, Michigan State University, East Lansing, Michigan, United States of America, DOE-Great Lake Bioenergy Research Center, Michigan State University, East Lansing, Michigan, United States of America
Roles Conceptualization, Investigation, Project administration, Supervision, Writing – review & editing
Affiliation Department of Plant Biology, Michigan State University, East Lansing, Michigan, United States of America
- Shin-Han Shiu,
- Melissa D. Lehti-Shiu

- Published: May 23, 2024
- https://doi.org/10.1371/journal.pbio.3002612
- Peer Review
- Reader Comments
Scientific advances due to conceptual or technological innovations can be revealed by examining how research topics have evolved. But such topical evolution is difficult to uncover and quantify because of the large body of literature and the need for expert knowledge in a wide range of areas in a field. Using plant biology as an example, we used machine learning and language models to classify plant science citations into topics representing interconnected, evolving subfields. The changes in prevalence of topical records over the last 50 years reflect shifts in major research trends and recent radiation of new topics, as well as turnover of model species and vastly different plant science research trajectories among countries. Our approaches readily summarize the topical diversity and evolution of a scientific field with hundreds of thousands of relevant papers, and they can be applied broadly to other fields.
Citation: Shiu S-H, Lehti-Shiu MD (2024) Assessing the evolution of research topics in a biological field using plant science as an example. PLoS Biol 22(5): e3002612. https://doi.org/10.1371/journal.pbio.3002612
Academic Editor: Ulrich Dirnagl, Charite Universitatsmedizin Berlin, GERMANY
Received: October 16, 2023; Accepted: April 4, 2024; Published: May 23, 2024
Copyright: © 2024 Shiu, Lehti-Shiu. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: The plant science corpus data are available through Zenodo ( https://zenodo.org/records/10022686 ). The codes for the entire project are available through GitHub ( https://github.com/ShiuLab/plant_sci_hist ) and Zenodo ( https://doi.org/10.5281/zenodo.10894387 ).
Funding: This work was supported by the National Science Foundation (IOS-2107215 and MCB-2210431 to MDL and SHS; DGE-1828149 and IOS-2218206 to SHS), Department of Energy grant Great Lakes Bioenergy Research Center (DE-SC0018409 to SHS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests: The authors have declared that no competing interests exist.
Abbreviations: BERT, Bidirectional Encoder Representations from Transformers; br, brassinosteroid; ccTLD, country code Top Level Domain; c-Tf-Idf, class-based Tf-Idf; ChatGPT, Chat Generative Pretrained Transformer; ga, gibberellic acid; LOWESS, locally weighted scatterplot smoothing; MeSH, Medical Subject Heading; SHAP, SHapley Additive exPlanations; SJR, SCImago Journal Rank; Tf-Idf, Term frequency-Inverse document frequency; UMAP, Uniform Manifold Approximation and Projection
Introduction
The explosive growth of scientific data in recent years has been accompanied by a rapidly increasing volume of literature. These records represent a major component of our scientific knowledge and embody the history of conceptual and technological advances in various fields over time. Our ability to wade through these records is important for identifying relevant literature for specific topics, a crucial practice of any scientific pursuit [ 1 ]. Classifying the large body of literature into topics can provide a useful means to identify relevant literature. In addition, these topics offer an opportunity to assess how scientific fields have evolved and when major shifts in took place. However, such classification is challenging because the relevant articles in any topic or domain can number in the tens or hundreds of thousands, and the literature is in the form of natural language, which takes substantial effort and expertise to process [ 2 , 3 ]. In addition, even if one could digest all literature in a field, it would still be difficult to quantify such knowledge.
In the last several years, there has been a quantum leap in natural language processing approaches due to the feasibility of building complex deep learning models with highly flexible architectures [ 4 , 5 ]. The development of large language models such as Bidirectional Encoder Representations from Transformers (BERT; [ 6 ]) and Chat Generative Pretrained Transformer (ChatGPT; [ 7 ]) has enabled the analysis, generation, and modeling of natural language texts in a wide range of applications. The success of these applications is, in large part, due to the feasibility of considering how the same words are used in different contexts when modeling natural language [ 6 ]. One such application is topic modeling, the practice of establishing statistical models of semantic structures underlying a document collection. Topic modeling has been proposed for identifying scientific hot topics over time [ 1 ], for example, in synthetic biology [ 8 ], and it has also been applied to, for example, automatically identify topical scenes in images [ 9 ] and social network topics [ 10 ], discover gene programs highly correlated with cancer prognosis [ 11 ], capture “chromatin topics” that define cell-type differences [ 12 ], and investigate relationships between genetic variants and disease risk [ 13 ]. Here, we use topic modeling to ask how research topics in a scientific field have evolved and what major changes in the research trends have taken place, using plant science as an example.
Plant science corpora allow classification of major research topics
Plant science, broadly defined, is the study of photosynthetic species, their interactions with biotic/abiotic environments, and their applications. For modeling plant science topical evolution, we first identified a collection of plant science documents (i.e., corpus) using a text classification approach. To this end, we first collected over 30 million PubMed records and narrowed down candidate plant science records by searching for those with plant-related terms and taxon names (see Materials and methods ). Because there remained a substantial number of false positives (i.e., biomedical records mentioning plants in passing), a set of positive plant science examples from the 17 plant science journals with the highest numbers of plant science publications covering a wide range of subfields and a set of negative examples from journals with few candidate plant science records were used to train 4 types of text classification models (see Materials and methods ). The best text classification model performed well (F1 = 0.96, F1 of a naïve model = 0.5, perfect model = 1) where the positive and negative examples were clearly separated from each other based on prediction probability of the hold-out testing dataset (false negative rate = 2.6%, false positive rate = 5.2%, S1A and S1B Fig ). The false prediction rate for documents from the 17 plant science journals annotated with the Medical Subject Heading (MeSH) term “Plants” in NCBI was 11.7% (see Materials and methods ). The prediction probability distribution of positive instances with the MeSH term has an expected left-skew to lower values ( S1C Fig ) compared with the distributions of all positive instances ( S1A Fig ). Thus, this subset with the MeSH term is a skewed representation of articles from these 17 major plant science journals. To further benchmark the validity of the plant science records, we also conducted manual annotation of 100 records where the false positive and false negative rates were 14.6% and 10.6%, respectively (see Materials and methods ). Using 12 other plant science journals not included as positive examples as benchmarks, the false negative rate was 9.9% (see Materials and methods ). Considering the range of false prediction rate estimates with different benchmarks, we should emphasize that the model built with the top 17 plant science journals represents a substantial fraction of plant science publications but with biases. Applying the model to the candidate plant science record led to 421,658 positive predictions, hereafter referred to as “plant science records” ( S1D Fig and S1 Data ).
To better understand how the models classified plant science articles, we identified important terms from a more easily interpretable model (Term frequency-Inverse document frequency (Tf-Idf) model; F1 = 0.934) using Shapley Additive Explanations [ 14 ]; 136 terms contributed to predicting plant science records (e.g., Arabidopsis, xylem, seedling) and 138 terms contributed to non-plant science record predictions (e.g., patients, clinical, mice; Tf-Idf feature sheet, S1 Data ). Plant science records as well as PubMed articles grew exponentially from 1950 to 2020 ( Fig 1A ), highlighting the challenges of digesting the rapidly expanding literature. We used the plant science records to perform topic modeling, which consisted of 4 steps: representing each record as a BERT embedding, reducing dimensionality, clustering, and identifying the top terms by calculating class (i.e., topic)-based Tf-Idf (c-Tf-Idf; [ 15 ]). The c-Tf-Idf represents the frequency of a term in the context of how rare the term is to reduce the influence of common words. SciBERT [ 16 ] was the best model among those tested ( S2 Data ) and was used for building the final topic model, which classified 372,430 (88.3%) records into 90 topics defined by distinct combinations of terms ( S3 Data ). The topics contained 620 to 16,183 records and were named after the top 4 to 5 terms defining the topical areas ( Fig 1B and S3 Data ). For example, the top 5 terms representing the largest topic, topic 61 (16,183 records), are “qtl,” “resistance,” “wheat,” “markers,” and “traits,” which represent crop improvement studies using quantitative genetics.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
(A) Numbers of PubMed (magenta) and plant science (green) records between 1950 and 2020. (a, b, c) Coefficients of the exponential function, y = ae b . Data for the plot are in S1 Data . (B) Numbers of documents for the top 30 plant science topics. Each topic is designated by an index number (left) and the top 4–6 terms with the highest cTf-Idf values (right). Data for the plot are in S3 Data . (C) Two-dimensional representation of the relationships between plant science records generated by Uniform Manifold Approximation and Projection (UMAP, [ 17 ]) using SciBERT embeddings of plant science records. All topics panel: Different topics are assigned different colors. Outlier panel: UMAP representation of all records (gray) with outlier records in red. Blue dotted circles: areas with relatively high densities indicating topics that are below the threshold for inclusion in a topic. In the 8 UMAP representations on the right, records for example topics are in red and the remaining records in gray. Blue dotted circles indicate the relative position of topic 48.
https://doi.org/10.1371/journal.pbio.3002612.g001
Records with assigned topics clustered into distinct areas in a two-dimensional (2D) space ( Fig 1C , for all topics, see S4 Data ). The remaining 49,228 outlier records not assigned to any topic (11.7%, middle panel, Fig 1C ) have 3 potential sources. First, some outliers likely belong to unique topics but have fewer records than the threshold (>500, blue dotted circles, Fig 1C ). Second, some of the many outliers dispersed within the 2D space ( Fig 1C ) were not assigned to any single topic because they had relatively high prediction scores for multiple topics ( S2 Fig ). These likely represent studies across subdisciplines in plant science. Third, some outliers are likely interdisciplinary studies between plant science and other domains, such as chemistry, mathematics, and physics. Such connections can only be revealed if records from other domains are included in the analyses.
Topical clusters reveal closely related topics but with distinct key term usage
Related topics tend to be located close together in the 2D representation (e.g., topics 48 and 49, Fig 1C ). We further assessed intertopical relationships by determining the cosine similarities between topics using cTf-Idfs ( Figs 2A and S3 ). In this topic network, some topics are closely related and form topic clusters. For example, topics 25, 26, and 27 collectively represent a more general topic related to the field of plant development (cluster a , lower left in Fig 2A ). Other topic clusters represent studies of stress, ion transport, and heavy metals ( b ); photosynthesis, water, and UV-B ( c ); population and community biology (d); genomics, genetic mapping, and phylogenetics ( e , upper right); and enzyme biochemistry ( f , upper left in Fig 2A ).
(A) Graph depicting the degrees of similarity (edges) between topics (nodes). Between each topic pair, a cosine similarity value was calculated using the cTf-Idf values of all terms. A threshold similarity of 0.6 was applied to illustrate the most related topics. For the full matrix presented as a heatmap, see S4 Fig . The nodes are labeled with topic index numbers and the top 4–6 terms. The colors and width of the edges are defined based on cosine similarity. Example topic clusters are highlighted in yellow and labeled a through f (blue boxes). (B, C) Relationships between the cTf-Idf values (see S3 Data ) of the top terms for topics 26 and 27 (B) and for topics 25 and 27 (C) . Only terms with cTf-Idf ≥ 0.6 are labeled. Terms with cTf-Idf values beyond the x and y axis limit are indicated by pink arrows and cTf-Idf values. (D) The 2D representation in Fig 1C is partitioned into graphs for different years, and example plots for every 5-year period since 1975 are shown. Example topics discussed in the text are indicated. Blue arrows connect the areas occupied by records of example topics across time periods to indicate changes in document frequencies.
https://doi.org/10.1371/journal.pbio.3002612.g002
Topics differed in how well they were connected to each other, reflecting how general the research interests or needs are (see Materials and methods ). For example, topic 24 (stress mechanisms) is the most well connected with median cosine similarity = 0.36, potentially because researchers in many subfields consider aspects of plant stress even though it is not the focus. The least connected topics include topic 21 (clock biology, 0.12), which is surprising because of the importance of clocks in essentially all aspects of plant biology [ 18 ]. This may be attributed, in part, to the relatively recent attention in this area.
Examining topical relationships and the cTf-Idf values of terms also revealed how related topics differ. For example, topic 26 is closely related to topics 27 and 25 (cluster a on the lower left of Fig 2A ). Topics 26 and 27 both contain records of developmental process studies mainly in Arabidopsis ( Fig 2B ); however, topic 26 is focused on the impact of light, photoreceptors, and hormones such as gibberellic acids (ga) and brassinosteroids (br), whereas topic 27 is focused on flowering and floral development. Topic 25 is also focused on plant development but differs from topic 27 because it contains records of studies mainly focusing on signaling and auxin with less emphasis on Arabidopsis ( Fig 2C ). These examples also highlight the importance of using multiple top terms to represent the topics. The similarities in cTf-Idfs between topics were also useful for measuring the editorial scope (i.e., diverse, or narrow) of journals publishing plant science papers using a relative topic diversity measure (see Materials and methods ). For example, Proceedings of the National Academy of Sciences , USA has the highest diversity, while Theoretical and Applied Genetics has the lowest ( S4 Fig ). One surprise is the relatively low diversity of American Journal of Botany , which focuses on plant ecology, systematics, development, and genetics. The low diversity is likely due to the relatively larger number of cellular and molecular science records in PubMed, consistent with the identification of relatively few topical areas relevant to studies at the organismal, population, community, and ecosystem levels.
Investigation of the relative prevalence of topics over time reveals topical succession
We next asked whether relationships between topics reflect chronological progression of certain subfields. To address this, we assessed how prevalent topics were over time using dynamic topic modeling [ 19 ]. As shown in Fig 2D , there is substantial fluctuation in where the records are in the 2D space over time. For example, topic 44 (light, leaves, co, synthesis, photosynthesis) is among the topics that existed in 1975 but has diminished gradually since. In 1985, topic 39 (Agrobacterium-based transformation) became dense enough to be visualized. Additional examples include topics 79 (soil heavy metals), 42 (differential expression), and 82 (bacterial community metagenomics), which became prominent in approximately 2005, 2010, and 2020, respectively ( Fig 2D ). In addition, animating the document occupancy in the 2D space over time revealed a broad change in patterns over time: Some initially dense areas became sparse over time and a large number of topics in areas previously only loosely occupied at the turn of the century increased over time ( S5 Data ).
While the 2D representations reveal substantial details on the evolution of topics, comparison over time is challenging because the number of plant science records has grown exponentially ( Fig 1A ). To address this, the records were divided into 50 chronological bins each with approximately 8,400 records to make cross-bin comparisons feasible ( S6 Data ). We should emphasize that, because of the way the chronological bins were split, the number of records for each topic in each bin should be treated as a normalized value relative to all other topics during the same period. Examining this relative prevalence of topics across bins revealed a clear pattern of topic succession over time (one topic evolved into another) and the presence of 5 topical categories ( Fig 3 ). The topics were categorized based on their locally weighted scatterplot smoothing (LOWESS) fits and ordered according to timing of peak frequency ( S7 and S8 Data , see Materials and methods ). In Fig 3 , the relative decrease in document frequency does not mean that research output in a topic is dwindling. Because each row in the heatmap is normalized based on the minimum and maximum values within each topic, there still can be substantial research output in terms of numbers of publications even when the relative frequency is near zero. Thus, a reduced relative frequency of a topic reflects only a below-average growth rate compared with other topical areas.
(A-E) A heat map of relative topic frequency over time reveals 5 topical categories: (A) stable, (B) early, (C) transitional, (D) sigmoidal, and (E) rising. The x axis denotes different time bins with each bin containing a similar number of documents to account for the exponential growth of plant science records over time. The sizes of all bins except the first are drawn to scale based on the beginning and end dates. The y axis lists different topics denoted by the label and top 4 to 5 terms. In each cell, the prevalence of a topic in a time bin is colored according to the min-max normalized cTf-Idf values for that topic. Light blue dotted lines delineate different decades. The arrows left of a subset of topic labels indicate example relationships between topics in topic clusters. Blue boxes with labels a–f indicate topic clusters, which are the same as those in Fig 2 . Connecting lines indicate successional trends. Yellow circles/lines 1 – 3: 3 major transition patterns. The original data are in S5 Data .
https://doi.org/10.1371/journal.pbio.3002612.g003
The first topical category is a stable category with 7 topics mostly established before the 1980s that have since remained stable in terms of prevalence in the plant science records (top of Fig 3A ). These topics represent long-standing plant science research foci, including studies of plant physiology (topics 4, 58, and 81), genetics (topic 61), and medicinal plants (topic 53). The second category contains 8 topics established before the 1980s that have mostly decreased in prevalence since (the early category, Fig 3B ). Two examples are physiological and morphological studies of hormone action (topic 45, the second in the early category) and the characterization of protein, DNA, and RNA (topic 18, the second to last). Unlike other early topics, topic 78 (paleobotany and plant evolution studies, the last topic in Fig 3B ) experienced a resurgence in the early 2000s due to the development of new approaches and databases and changes in research foci [ 20 ].
The 33 topics in the third, transitional category became prominent in the 1980s, 1990s, or even 2000s but have clearly decreased in prevalence ( Fig 3C ). In some cases, the early and the transitional topics became less prevalent because of topical succession—refocusing of earlier topics led to newer ones that either show no clear sign of decrease (the sigmoidal category, Fig 3D ) or continue to increase in prevalence (the rising category, Fig 3E ). Consistent with the notion of topical succession, topics within each topic cluster ( Fig 2 ) were found across topic categories and/or were prominent at different time periods (indicated by colored lines linking topics, Fig 3 ). One example is topics in topic cluster b (connected with light green lines and arrows, compare Figs 2 and 3 ); the study of cation transport (topic 47, the third in the transitional category), prominent in the 1980s and early 1990s, is connected to 5 other topics, namely, another transitional topic 29 (cation channels and their expression) peaking in the 2000s and early 2010s, sigmoidal topics 24 and 28 (stress response, tolerance mechanisms) and 30 (heavy metal transport), which rose to prominence in mid-2000s, and the rising topic 42 (stress transcriptomic studies), which increased in prevalence in the mid-2010s.
The rise and fall of topics can be due to a combination of technological or conceptual breakthroughs, maturity of the field, funding constraints, or publicity. The study of transposable elements (topic 62) illustrates the effect of publicity; the rise in this field coincided with Barbara McClintock’s 1983 Nobel Prize but not with the publication of her studies in the 1950s [ 21 ]. The reduced prevalence in early 2000 likely occurred in part because analysis of transposons became a central component of genome sequencing and annotation studies, rather than dedicated studies. In addition, this example indicates that our approaches, while capable of capturing topical trends, cannot be used to directly infer major papers leading to the growth of a topic.
Three major topical transition patterns signify shifts in research trends
Beyond the succession of specific topics, 3 major transitions in the dynamic topic graph should be emphasized: (1) the relative decreasing trend of early topics in the late 1970s and early 1980s; (2) the rise of transitional topics in late 1980s; and (3) the relative decreasing trend of transitional topics in the late 1990s and early 2000s, which coincided with a radiation of sigmoidal and rising topics (yellow circles, Fig 3 ). The large numbers of topics involved in these transitions suggest major shifts in plant science research. In transition 1, early topics decreased in relative prevalence in the late 1970s to early 1980s, which coincided with the rise of transitional topics over the following decades (circle 1, Fig 3 ). For example, there was a shift from the study of purified proteins such as enzymes (early topic 48, S5A Fig ) to molecular genetic dissection of genes, proteins, and RNA (transitional topic 35, S5B Fig ) enabled by the wider adoption of recombinant DNA and molecular cloning technologies in late 1970s [ 22 ]. Transition 2 (circle 2, Fig 3 ) can be explained by the following breakthroughs in the late 1980s: better approaches to create transgenic plants and insertional mutants [ 23 ], more efficient creation of mutant plant libraries through chemical mutagenesis (e.g., [ 24 ]), and availability of gene reporter systems such as β-glucuronidase [ 25 ]. Because of these breakthroughs, molecular genetics studies shifted away from understanding the basic machinery to understanding the molecular underpinnings of specific processes, such as molecular mechanisms of flower and meristem development and the action of hormones such as auxin (topic 27, S5C Fig ); this type of research was discussed as a future trend in 1988 [ 26 ] and remains prevalent to this date. Another example is gene silencing (topic 12), which became a focal area of study along with the widespread use of transgenic plants [ 27 ].
Transition 3 is the most drastic: A large number of transitional, sigmoidal, and rising topics became prevalent nearly simultaneously at the turn of the century (circle 3, Fig 3 ). This period also coincides with a rapid increase in plant science citations ( Fig 1A ). The most notable breakthroughs included the availability of the first plant genome in 2000 [ 28 ], increasing ease and reduced cost of high-throughput sequencing [ 29 ], development of new mass spectrometry–based platforms for analyzing proteins [ 30 ], and advancements in microscopic and optical imaging approaches [ 31 ]. Advances in genomics and omics technology also led to an increase in stress transcriptomics studies (42, S5D Fig ) as well as studies in many other topics such as epigenetics (topic 11), noncoding RNA analysis (13), genomics and phylogenetics (80), breeding (41), genome sequencing and assembly (60), gene family analysis (23), and metagenomics (82 and 55).
In addition to the 3 major transitions across all topics, there were also transitions within topics revealed by examining the top terms for different time bins (heatmaps, S5 Fig ). Taken together, these observations demonstrate that knowledge about topical evolution can be readily revealed through topic modeling. Such knowledge is typically only available to experts in specific areas and is difficult to summarize manually, as no researcher has a command of the entire plant science literature.
Analysis of taxa studied reveals changes in research trends
Changes in research trends can also be illustrated by examining changes in the taxa being studied over time ( S9 Data ). There is a strong bias in the taxa studied, with the record dominated by research models and economically important taxa ( S6 Fig ). Flowering plants (Magnoliopsida) are found in 93% of records ( S6A Fig ), and the mustard family Brassicaceae dominates at the family level ( S6B Fig ) because the genus Arabidopsis contributes to 13% of plant science records ( Fig 4A ). When examining the prevalence of taxa being studied over time, clear patterns of turnover emerged similar to topical succession ( Figs 4B , S6C, and S6D ; Materials and methods ). Given that Arabidopsis is mentioned in more publications than other species we analyzed, we further examined the trends for Arabidopsis publications. The increase in the normalized number (i.e., relative to the entire plant science corpus) of Arabidopsis records coincided with advocacy of its use as a model system in the late 1980s [ 32 ]. While it remains a major plant model, there has been a decrease in overall Arabidopsis publications relative to all other plant science publications since 2011 (blue line, normalized total, Fig 4C ). Because the same chronological bins, each with same numbers of records, from the topic-over-time analysis ( Fig 3 ) were used, the decrease here does not mean that there were fewer Arabidopsis publications—in fact, the number of Arabidopsis papers has remained steady since 2011. This decrease means that Arabidopsis-related publications represent a relatively smaller proportion of plant science records. Interestingly, this decrease took place much earlier (approximately 2005) and was steeper in the United States (red line, Fig 4C ) than in all countries combined (blue line, Fig 4C ).
(A) Percentage of records mentioning specific genera. (B) Change in the prevalence of genera in plant science records over time. (C) Changes in the normalized numbers of all records (blue) and records from the US (red) mentioning Arabidopsis over time. The lines are LOWESS fits with fraction parameter = 0.2. (D) Topical over (red) and under (blue) representation among 5 genera with the most plant science records. LLR: log 2 likelihood ratios of each topic in each genus. Gray: topic-species combination not significantly enriched at the 5% level based on enrichment p -values adjusted for multiple testing with the Benjamini–Hochberg method [ 33 ]. The data used for plotting are in S9 Data . The statistics for all topics are in S10 Data .
https://doi.org/10.1371/journal.pbio.3002612.g004
Assuming that the normalized number of publications reflects the relative intensity of research activities, one hypothesis for the relative decrease in focus on Arabidopsis is that advances in, for example, plant transformation, genetic manipulation, and genome research have allowed the adoption of more previously nonmodel taxa. Consistent with this, there was a precipitous increase in the number of genera being published in the mid-90s to early 2000s during which approaches for plant transgenics became established [ 34 ], but the number has remained steady since then ( S7A Fig ). The decrease in the proportion of Arabidopsis papers is also negatively correlated with the timing of an increase in the number of draft genomes ( S7B Fig and S9 Data ). It is plausible that genome availability for other species may have contributed to a shift away from Arabidopsis. Strikingly, when we analyzed US National Science Foundation records, we found that the numbers of funded grants mentioning Arabidopsis ( S7C Fig ) have risen and fallen in near perfect synchrony with the normalized number of Arabidopsis publication records (red line, Fig 4C ). This finding likely illustrates the impact of funding on Arabidopsis research.
By considering both taxa information and research topics, we can identify clear differences in the topical areas preferred by researchers using different plant taxa ( Fig 4D and S10 Data ). For example, studies of auxin/light signaling, the circadian clock, and flowering tend to be carried out in Arabidopsis, while quantitative genetic studies of disease resistance tend to be done in wheat and rice, glyphosate research in soybean, and RNA virus research in tobacco. Taken together, joint analyses of topics and species revealed additional details about changes in preferred models over time, and the preferred topical areas for different taxa.
Countries differ in their contributions to plant science and topical preference
We next investigated whether there were geographical differences in topical preference among countries by inferring country information from 330,187 records (see Materials and methods ). The 10 countries with the most records account for 73% of the total, with China and the US contributing to approximately 18% each ( Fig 5A ). The exponential growth in plant science records (green line, Fig 1A ) was in large part due to the rapid rise in annual record numbers in China and India ( Fig 5B ). When we examined the publication growth rates using the top 17 plant science journals, the general patterns remained the same ( S7D Fig ). On the other hand, the US, Japan, Germany, France, and Great Britain had slower rates of growth compared with all non-top 10 countries. The rapid increase in records from China and India was accompanied by a rapid increase in metrics measuring journal impact ( Figs 5C and S8 and S9 Data ). For example, using citation score ( Fig 5C , see Materials and methods ), we found that during a 22-year period China (dark green) and India (light green) rapidly approached the global average (y = 0, yellow), whereas some of the other top 10 countries, particularly the US (red) and Japan (yellow green), showed signs of decrease ( Fig 5C ). It remains to be determined whether these geographical trends reflect changes in priority, investment, and/or interest in plant science research.
(A) Numbers of plant science records for countries with the 10 highest numbers. (B) Percentage of all records from each of the top 10 countries from 1980 to 2020. (C) Difference in citation scores from 1999 to 2020 for the top 10 countries. (D) Shown for each country is the relationship between the citation scores averaged from 1999 to 2020 and the slope of linear fit with year as the predictive variable and citation score as the response variable. The countries with >400 records and with <10% missing impact values are included. Data used for plots (A–D) are in S11 Data . (E) Correlation in topic enrichment scores between the top 10 countries. PCC, Pearson’s correlation coefficient, positive in red, negative in blue. Yellow rectangle: countries with more similar topical preferences. (F) Enrichment scores (LLR, log likelihood ratio) of selected topics among the top 10 countries. Red: overrepresentation, blue: underrepresentation. Gray: topic-country combination that is not significantly enriched at the 5% level based on enrichment p -values adjusted for multiple testing with the Benjamini–Hochberg method (for all topics and plotting data, see S12 Data ).
https://doi.org/10.1371/journal.pbio.3002612.g005
Interestingly, the relative growth/decline in citation scores over time (measured as the slope of linear fit of year versus citation score) was significantly and negatively correlated with average citation score ( Fig 5D ); i.e., countries with lower overall metrics tended to experience the strongest increase in citation scores over time. Thus, countries that did not originally have a strong influence on plant sciences now have increased impact. These patterns were also observed when using H-index or journal rank as metrics ( S8 Fig and S11 Data ) and were not due to increased publication volume, as the metrics were normalized against numbers of records from each country (see Materials and methods ). In addition, the fact that different metrics with different caveats and assumptions yielded consistent conclusions indicates the robustness of our observations. We hypothesize that this may be a consequence of the ease in scientific communication among geographically isolated research groups. It could also be because of the prevalence of online journals that are open access, which makes scientific information more readily accessible. Or it can be due to the increasing international collaboration. In any case, the causes for such regression toward the mean are not immediately clear and should be addressed in future studies.
We also assessed how the plant research foci of countries differ by comparing topical preference (i.e., the degree of enrichment of plant science records in different topics) between countries. For example, Italy and Spain cluster together (yellow rectangle, Fig 5E ) partly because of similar research focusing on allergens (topic 0) and mycotoxins (topic 54) and less emphasis on gene family (topic 23) and stress tolerance (topic 28) studies ( Fig 5F , for the fold enrichment and corrected p -values of all topics, see S12 Data ). There are substantial differences in topical focus between countries ( S9 Fig ). For example, research on new plant compounds associated with herbal medicine (topic 69) is a focus in China but not in the US, but the opposite is true for population genetics and evolution (topic 86) ( Fig 5F ). In addition to revealing how plant science research has evolved over time, topic modeling provides additional insights into differences in research foci among different countries, which are informative for science policy considerations.
In this study, topic modeling revealed clear transitions among research topics, which represent shifts in research trends in plant sciences. One limitation of our study is the bias in the PubMed-based corpus. The cellular, molecular, and physiological aspects of plant sciences are well represented, but there are many fewer records related to evolution, ecology, and systematics. Our use of titles/abstracts from the top 17 plant science journals as positive examples allowed us to identify papers we typically see in these journals, but this may have led to us missing “outlier” articles, which may be the most exciting. Another limitation is the need to assign only one topic to a record when a study is interdisciplinary and straddles multiple topics. Furthermore, a limited number of large, inherently heterogeneous topics were summarized to provide a more concise interpretation, which undoubtedly underrepresents the diversity of plant science research. Despite these limitations, dynamic topic modeling revealed changes in plant science research trends that coincide with major shifts in biological science. While we were interested in identifying conceptual advances, our approach can identify the trend but the underlying causes for such trends, particularly key records leading to the growth in certain topics, still need to be identified. It also remains to be determined which changes in research trends lead to paradigm shifts as defined by Kuhn [ 35 ].
The key terms defining the topics frequently describe various technologies (e.g., topic 38/39: transformation, 40: genome editing, 59: genetic markers, 65: mass spectrometry, 69: nuclear magnetic resonance) or are indicative of studies enabled through molecular genetics and omics technologies (e.g., topic 8/60: genome, 11: epigenetic modifications, 18: molecular biological studies of macromolecules, 13: small RNAs, 61: quantitative genetics, 82/84: metagenomics). Thus, this analysis highlights how technological innovation, particularly in the realm of omics, has contributed to a substantial number of research topics in the plant sciences, a finding that likely holds for other scientific disciplines. We also found that the pattern of topic evolution is similar to that of succession, where older topics have mostly decreased in relative prevalence but appear to have been superseded by newer ones. One example is the rise of transcriptome-related topics and the correlated, reduced focus on regulation at levels other than transcription. This raises the question of whether research driven by technology negatively impacts other areas of research where high-throughput studies remain challenging.
One observation on the overall trends in plant science research is the approximately 10-year cycle in major shifts. One hypothesis is related to not only scientific advances but also to the fashion-driven aspect of science. Nonetheless, given that there were only 3 major shifts and the sample size is small, it is difficult to speculate as to why they happened. By analyzing the country of origin, we found that China and India have been the 2 major contributors to the growth in the plant science records in the last 20 years. Our findings also show an equalizing trend in global plant science where countries without a strong plant science publication presence have had an increased impact over the last 20 years. In addition, we identified significant differences in research topics between countries reflecting potential differences in investment and priorities. Such information is important for discerning differences in research trends across countries and can be considered when making policy decisions about research directions.
Materials and methods
Collection and preprocessing of a candidate plant science corpus.
For reproducibility purposes, a random state value of 20220609 was used throughout the study. The PubMed baseline files containing citation information ( ftp://ftp.ncbi.nlm.nih.gov/pubmed/baseline/ ) were downloaded on November 11, 2021. To narrow down the records to plant science-related citations, a candidate citation was identified as having, within the titles and/or abstracts, at least one of the following words: “plant,” “plants,” “botany,” “botanical,” “planta,” and “plantarum” (and their corresponding upper case and plural forms), or plant taxon identifiers from NCBI Taxonomy ( https://www.ncbi.nlm.nih.gov/taxonomy ) or USDA PLANTS Database ( https://plants.sc.egov.usda.gov/home ). Note the search terms used here have nothing to do with the values of the keyword field in PubMed records. The taxon identifiers include all taxon names including and at taxonomic levels below “Viridiplantae” till the genus level (species names not used). This led to 51,395 search terms. After looking for the search terms, qualified entries were removed if they were duplicated, lacked titles and/or abstracts, or were corrections, errata, or withdrawn articles. This left 1,385,417 citations, which were considered the candidate plant science corpus (i.e., a collection of texts). For further analysis, the title and abstract for each citation were combined into a single entry. Text was preprocessed by lowercasing, removing stop-words (i.e., common words), removing non-alphanumeric and non-white space characters (except Greek letters, dashes, and commas), and applying lemmatization (i.e., grouping inflected forms of a word as a single word) for comparison. Because lemmatization led to truncated scientific terms, it was not included in the final preprocessing pipeline.
Definition of positive/negative examples
Upon closer examination, a large number of false positives were identified in the candidate plant science records. To further narrow down citations with a plant science focus, text classification was used to distinguish plant science and non-plant science articles (see next section). For the classification task, a negative set (i.e., non-plant science citations) was defined as entries from 7,360 journals that appeared <20 times in the filtered data (total = 43,329, journal candidate count, S1 Data ). For the positive examples (i.e., true plant science citations), 43,329 plant science citations (positive examples) were sampled from 17 established plant science journals each with >2,000 entries in the filtered dataset: “Plant physiology,” “Frontiers in plant science,” “Planta,” “The Plant journal: for cell and molecular biology,” “Journal of experimental botany,” “Plant molecular biology,” “The New phytologist,” “The Plant cell,” “Phytochemistry,” “Plant & cell physiology,” “American journal of botany,” “Annals of botany,” “BMC plant biology,” “Tree physiology,” “Molecular plant-microbe interactions: MPMI,” “Plant biology,” and “Plant biotechnology journal” (journal candidate count, S1 Data ). Plant biotechnology journal was included, but only 1,894 records remained after removal of duplicates, articles with missing info, and/or withdrawn articles. The positive and negative sets were randomly split into training and testing subsets (4:1) while maintaining a 1:1 positive-to-negative ratio.
Text classification based on Tf and Tf-Idf
Instead of using the preprocessed text as features for building classification models directly, text embeddings (i.e., representations of texts in vectors) were used as features. These embeddings were generated using 4 approaches (model summary, S1 Data ): Term-frequency (Tf), Tf-Idf [ 36 ], Word2Vec [ 37 ], and BERT [ 6 ]. The Tf- and Tf-Idf-based features were generated with CountVectorizer and TfidfVectorizer, respectively, from Scikit-Learn [ 38 ]. Different maximum features (1e4 to 1e5) and n-gram ranges (uni-, bi-, and tri-grams) were tested. The features were selected based on the p- value of chi-squared tests testing whether a feature had a higher-than-expected value among the positive or negative classes. Four different p- value thresholds were tested for feature selection. The selected features were then used to retrain vectorizers with the preprocessed training texts to generate feature values for classification. The classification model used was XGBoost [ 39 ] with 5 combinations of the following hyperparameters tested during 5-fold stratified cross-validation: min_child_weight = (1, 5, 10), gamma = (0.5, 1, 1.5, 2.5), subsample = (0.6, 0.8, 1.0), colsample_bytree = (0.6, 0.8, 1.0), and max_depth = (3, 4, 5). The rest of the hyperparameters were held constant: learning_rate = 0.2, n_estimators = 600, objective = binary:logistic. RandomizedSearchCV from Scikit-Learn was used for hyperparameter tuning and cross-validation with scoring = F1-score.
Because the Tf-Idf model had a relatively high model performance and was relatively easy to interpret (terms are frequency-based, instead of embedding-based like those generated by Word2Vec and BERT), the Tf-Idf model was selected as input to SHapley Additive exPlanations (SHAP; [ 14 ]) to assess the importance of terms. Because the Tf-Idf model was based on XGBoost, a tree-based algorithm, the TreeExplainer module in SHAP was used to determine a SHAP value for each entry in the training dataset for each Tf-Idf feature. The SHAP value indicates the degree to which a feature positively or negatively affects the underlying prediction. The importance of a Tf-Idf feature was calculated as the average SHAP value of that feature among all instances. Because a Tf-Idf feature is generated based on a specific term, the importance of the Tf-Idf feature indicates the importance of the associated term.
Text classification based on Word2Vec
The preprocessed texts were first split into train, validation, and test subsets (8:1:1). The texts in each subset were converted to 3 n-gram lists: a unigram list obtained by splitting tokens based on the space character, or bi- and tri-gram lists built with Gensim [ 40 ]. Each n-gram list of the training subset was next used to fit a Skip-gram Word2Vec model with vector_size = 300, window = 8, min_count = (5, 10, or 20), sg = 1, and epochs = 30. The Word2Vec model was used to generate word embeddings for train, validate, and test subsets. In the meantime, a tokenizer was trained with train subset unigrams using Tensorflow [ 41 ] and used to tokenize texts in each subset and turn each token into indices to use as features for training text classification models. To ensure all citations had the same number of features (500), longer texts were truncated, and shorter ones were zero-padded. A deep learning model was used to train a text classifier with an input layer the same size as the feature number, an attention layer incorporating embedding information for each feature, 2 bidirectional Long-Short-Term-Memory layers (15 units each), a dense layer (64 units), and a final, output layer with 2 units. During training, adam, accuracy, and sparse_categorical_crossentropy were used as the optimizer, evaluation metric, and loss function, respectively. The training process lasted 30 epochs with early stopping if validation loss did not improve in 5 epochs. An F1 score was calculated for each n-gram list and min_count parameter combination to select the best model (model summary, S1 Data ).
Text classification based on BERT models
Two pretrained models were used for BERT-based classification: DistilBERT (Hugging face repository [ 42 ] model name and version: distilbert-base-uncased [ 43 ]) and SciBERT (allenai/scibert-scivocab-uncased [ 16 ]). In both cases, tokenizers were retrained with the training data. BERT-based models had the following architecture: the token indices (512 values for each token) and associated masked values as input layers, pretrained BERT layer (512 × 768) excluding outputs, a 1D pooling layer (768 units), a dense layer (64 units), and an output layer (2 units). The rest of the training parameters were the same as those for Word2Vec-based models, except training lasted for 20 epochs. Cross-validation F1-scores for all models were compared and used to select the best model for each feature extraction method, hyperparameter combination, and modeling algorithm or architecture (model summary, S1 Data ). The best model was the Word2Vec-based model (min_count = 20, window = 8, ngram = 3), which was applied to the candidate plant science corpus to identify a set of plant science citations for further analysis. The candidate plant science records predicted as being in the positive class (421,658) by the model were collectively referred to as the “plant science corpus.”
Plant science record classification
In PubMed, 1,384,718 citations containing “plant” or any plant taxon names (from the phylum to genus level) were considered candidate plant science citations. To further distinguish plant science citations from those in other fields, text classification models were trained using titles and abstracts of positive examples consisting of citations from 17 plant science journals, each with >2,000 entries in PubMed, and negative examples consisting of records from journals with fewer than 20 entries in the candidate set. Among 4 models tested the best model (built with Word2Vec embeddings) had a cross validation F1 of 0.964 (random guess F1 = 0.5, perfect model F1 = 1, S1 Data ). When testing the model using 17,330 testing set citations independent from the training set, the F1 remained high at 0.961.
We also conducted another analysis attempting to use the MeSH term “Plants” as a benchmark. Records with the MeSH term “Plants” also include pharmaceutical studies of plants and plant metabolites or immunological studies of plants as allergens in journals that are not generally considered plant science journals (e.g., Acta astronautica , International journal for parasitology , Journal of chromatography ) or journals from local scientific societies (e.g., Acta pharmaceutica Hungarica , Huan jing ke xue , Izvestiia Akademii nauk . Seriia biologicheskaia ). Because we explicitly labeled papers from such journals as negative examples, we focused on 4,004 records with the “Plants” MeSH term published in the 17 plant science journals that were used as positive instances and found that 88.3% were predicted as the positive class. Thus, based on the MeSH term, there is an 11.7% false prediction rate.
We also enlisted 5 plant science colleagues (3 advanced graduate students in plant biology and genetic/genome science graduate programs, 1 postdoctoral breeder/quantitative biologist, and 1 postdoctoral biochemist/geneticist) to annotate 100 randomly selected abstracts as a reviewer suggested. Each record was annotated by 2 colleagues. Among 85 entries where the annotations are consistent between annotators, 48 were annotated as negative but with 7 predicted as positive (false positive rate = 14.6%) and 37 were annotated as positive but with 4 predicted as negative (false negative rate = 10.8%). To further benchmark the performance of the text classification model, we identified another 12 journals that focus on plant science studies to use as benchmarks: Current opinion in plant biology (number of articles: 1,806), Trends in plant science (1,723), Functional plant biology (1,717), Molecular plant pathology (1,573), Molecular plant (1,141), Journal of integrative plant biology (1,092), Journal of plant research (1,032), Physiology and molecular biology of plants (830), Nature plants (538), The plant pathology journal (443). Annual review of plant biology (417), and The plant genome (321). Among the 12,611 candidate plant science records, 11,386 were predicted as positive. Thus, there is a 9.9% false negative rate.
Global topic modeling
BERTopic [ 15 ] was used for preliminary topic modeling with n-grams = (1,2) and with an embedding initially generated by DistilBERT, SciBERT, or BioBERT (dmis-lab/biobert-base-cased-v1.2; [ 44 ]). The embedding models converted preprocessed texts to embeddings. The topics generated based on the 3 embeddings were similar ( S2 Data ). However, SciBERT-, BioBERT-, and distilBERT-based embedding models had different numbers of outlier records (268,848, 293,790, and 323,876, respectively) with topic index = −1. In addition to generating the fewest outliers, the SciBERT-based model led to the highest number of topics. Therefore, SciBERT was chosen as the embedding model for the final round of topic modeling. Modeling consisted of 3 steps. First, document embeddings were generated with SentenceTransformer [ 45 ]. Second, a clustering model to aggregate documents into clusters using hdbscan [ 46 ] was initialized with min_cluster_size = 500, metric = euclidean, cluster_selection_method = eom, min_samples = 5. Third, the embedding and the initialized hdbscan model were used in BERTopic to model topics with neighbors = 10, nr_topics = 500, ngram_range = (1,2). Using these parameters, 90 topics were identified. The initial topic assignments were conservative, and 241,567 records were considered outliers (i.e., documents not assigned to any of the 90 topics). After assessing the prediction scores of all records generated from the fitted topic models, the 95-percentile score was 0.0155. This score was used as the threshold for assigning outliers to topics: If the maximum prediction score was above the threshold and this maximum score was for topic t , then the outlier was assigned to t . After the reassignment, 49,228 records remained outliers. To assess if some of the outliers were not assigned because they could be assigned to multiple topics, the prediction scores of the records were used to put records into 100 clusters using k- means. Each cluster was then assessed to determine if the outlier records in a cluster tended to have higher prediction scores across multiple topics ( S2 Fig ).
Topics that are most and least well connected to other topics
The most well-connected topics in the network include topic 24 (stress mechanisms, median cosine similarity = 0.36), topic 42 (genes, stress, and transcriptomes, 0.34), and topic 35 (molecular genetics, 0.32, all t test p -values < 1 × 10 −22 ). The least connected topics include topic 0 (allergen research, median cosine similarity = 0.12), topic 21 (clock biology, 0.12), topic 1 (tissue culture, 0.15), and topic 69 (identification of compounds with spectroscopic methods, 0.15; all t test p- values < 1 × 10 −24 ). Topics 0, 1, and 69 are specialized topics; it is surprising that topic 21 is not as well connected as explained in the main text.
Analysis of documents based on the topic model
Topical diversity among top journals with the most plant science records
Using a relative topic diversity measure (ranging from 0 to 10), we found that there was a wide range of topical diversity among 20 journals with the largest numbers of plant science records ( S3 Fig ). The 4 journals with the highest relative topical diversities are Proceedings of the National Academy of Sciences , USA (9.6), Scientific Reports (7.1), Plant Physiology (6.7), and PLOS ONE (6.4). The high diversities are consistent with the broad, editorial scopes of these journals. The 4 journals with the lowest diversities are American Journal of Botany (1.6), Oecologia (0.7), Plant Disease (0.7), and Theoretical and Applied Genetics (0.3), which reflects their discipline-specific focus and audience of classical botanists, ecologists, plant pathologists, and specific groups of geneticists.
Dynamic topic modeling
The codes for dynamic modeling were based on _topic_over_time.py in BERTopics and modified to allow additional outputs for debugging and graphing purposes. The plant science citations were binned into 50 subsets chronologically (for timestamps of bins, see S5 Data ). Because the numbers of documents increased exponentially over time, instead of dividing them based on equal-sized time intervals, which would result in fewer records at earlier time points and introduce bias, we divided them into time bins of similar size (approximately 8,400 documents). Thus, the earlier time subsets had larger time spans compared with later time subsets. If equal-size time intervals were used, the numbers of documents between the intervals would differ greatly; the earlier time points would have many fewer records, which may introduce bias. Prior to binning the subsets, the publication dates were converted to UNIX time (timestamp) in seconds; the plant science records start in 1917-11-1 (timestamp = −1646247600.0) and end in 2021-1-1 (timestamp = 1609477201). The starting dates and corresponding timestamps for the 50 subsets including the end date are in S6 Data . The input data included the preprocessed texts, topic assignments of records from global topic modeling, and the binned timestamps of records. Three additional parameters were set for topics_over_time, namely, nr_bin = 50 (number of bins), evolution_tuning = True, and global_tuning = False. The evolution_tuning parameter specified that averaged c-Tf-Idf values for a topic be calculated in neighboring time bins to reduce fluctuation in c-Tf-Idf values. The global_tuning parameter was set to False because of the possibility that some nonexisting terms could have a high c-Tf-Idf for a time bin simply because there was a high global c-Tf-Idf value for that term.
The binning strategy based on similar document numbers per bin allowed us to increase signal particularly for publications prior to the 90s. This strategy, however, may introduce more noise for bins with smaller time durations (i.e., more recent bins) because of publication frequencies (there can be seasonal differences in the number of papers published, biased toward, e.g., the beginning of the year or the beginning of a quarter). To address this, we examined the relative frequencies of each topic over time ( S7 Data ), but we found that recent time bins had similar variances in relative frequencies as other time bins. We also moderated the impact of variation using LOWESS (10% to 30% of the data points were used for fitting the trend lines) to determine topical trends for Fig 3 . Thus, the influence of the noise introduced via our binning strategy is expected to be minimal.
Topic categories and ordering
The topics were classified into 5 categories with contrasting trends: stable, early, transitional, sigmoidal, and rising. To define which category a topic belongs to, the frequency of documents over time bins for each topic was analyzed using 3 regression methods. We first tried 2 forecasting methods: recursive autoregressor (the ForecasterAutoreg class in the skforecast package) and autoregressive integrated moving average (ARIMA implemented in the pmdarima package). In both cases, the forecasting results did not clearly follow the expected trend lines, likely due to the low numbers of data points (relative frequency values), which resulted in the need to extensively impute missing data. Thus, as a third approach, we sought to fit the trendlines with the data points using LOWESS (implemented in the statsmodels package) and applied additional criteria for assigning topics to categories. When fitting with LOWESS, 3 fraction parameters (frac, the fraction of the data used when estimating each y-value) were evaluated (0.1, 0.2, 0.3). While frac = 0.3 had the smallest errors for most topics, in situations where there were outliers, frac = 0.2 or 0.1 was chosen to minimize mean squared errors ( S7 Data ).
The topics were classified into 5 categories based on the slopes of the fitted line over time: (1) stable: topics with near 0 slopes over time; (2) early: topics with negative (<−0.5) slopes throughout (with the exception of topic 78, which declined early on but bounced back by the late 1990s); (3) transitional: early positive (>0.5) slopes followed by negative slopes at later time points; (4) sigmoidal: early positive slopes followed by zero slopes at later time points; and (5) rising: continuously positive slopes. For each topic, the LOWESS fits were also used to determine when the relative document frequency reached its peak, first reaching a threshold of 0.6 (chosen after trial and error for a range of 0.3 to 0.9), and the overall trend. The topics were then ordered based on (1) whether they belonged to the stable category or not; (2) whether the trends were decreasing, stable, or increasing; (3) the time the relative document frequency first reached 0.6; and (4) the time that the overall peak was reached ( S8 Data ).
Taxa information
To identify a taxon or taxa in all plant science records, NCBI Taxonomy taxdump datasets were downloaded from the NCBI FTP site ( https://ftp.ncbi.nlm.nih.gov/pub/taxonomy/new_taxdump/ ) on September 20, 2022. The highest-level taxon was Viridiplantae, and all its child taxa were parsed and used as queries in searches against the plant science corpus. In addition, a species-over-time analysis was conducted using the same time bins as used for dynamic topic models. The number of records in different time bins for top taxa are in the genus, family, order, and additional species level sheet in S9 Data . The degree of over-/underrepresentation of a taxon X in a research topic T was assessed using the p -value of a Fisher’s exact test for a 2 × 2 table consisting of the numbers of records in both X and T, in X but not T, in T but not X, and in neither ( S10 Data ).
For analysis of plant taxa with genome information, genome data of taxa in Viridiplantae were obtained from the NCBI Genome data-hub ( https://www.ncbi.nlm.nih.gov/data-hub/genome ) on October 28, 2022. There were 2,384 plant genome assemblies belonging to 1,231 species in 559 genera (genome assembly sheet, S9 Data ). The date of the assembly was used as a proxy for the time when a genome was sequenced. However, some species have updated assemblies and have more recent data than when the genome first became available.
Taxa being studied in the plant science records
Flowering plants (Magnoliopsida) are found in 93% of records, while most other lineages are discussed in <1% of records, with conifers and related species being exceptions (Acrogynomsopermae, 3.5%, S6A Fig ). At the family level, the mustard (Brassicaceae), grass (Poaceae), pea (Fabaceae), and nightshade (Solanaceae) families are in 51% of records ( S6B Fig ). The prominence of the mustard family in plant science research is due to the Brassica and Arabidopsis genera ( Fig 4A ). When examining the prevalence of taxa being studied over time, clear patterns of turnovers emerged ( Figs 4B , S6C, and S6D ). While the study of monocot species (Liliopsida) has remained steady, there was a significant uptick in the prevalence of eudicot (eudicotyledon) records in the late 90s ( S6C Fig ), which can be attributed to the increased number of studies in the mustard, myrtle (Myrtaceae), and mint (Lamiaceae) families among others ( S6D Fig ). At the genus level, records mentioning Gossypium (cotton), Phaseolus (bean), Hordeum (wheat), and Zea (corn), similar to the topics in the early category, were prevalent till the 1980s or 1990s but have mostly decreased in number since ( Fig 4B ). In contrast, Capsicum , Arabidopsis , Oryza , Vitus , and Solanum research has become more prevalent over the last 20 years.
Geographical information for the plant science corpus
The geographical information (country) of authors in the plant science corpus was obtained from the address (AD) fields of first authors in Medline XML records accessible through the NCBI EUtility API ( https://www.ncbi.nlm.nih.gov/books/NBK25501/ ). Because only first author affiliations are available for records published before December 2014, only the first author’s location was considered to ensure consistency between records before and after that date. Among the 421,658 records in the plant science corpus, 421,585 had Medline records and 421,276 had unique PMIDs. Among the records with unique PMIDs, 401,807 contained address fields. For each of the remaining records, the AD field content was split into tokens with a “,” delimiter, and the token likely containing geographical info (referred to as location tokens) was selected as either the last token or the second to last token if the last token contained “@” indicating the presence of an email address. Because of the inconsistency in how geographical information was described in the location tokens (e.g., country, state, city, zip code, name of institution, and different combinations of the above), the following 4 approaches were used to convert location tokens into countries.
The first approach was a brute force search where full names and alpha-3 codes of current countries (ISO 3166–1), current country subregions (ISO 3166–2), and historical country (i.e., country that no longer exists, ISO 3166–3) were used to search the address fields. To reduce false positives using alpha-3 codes, a space prior to each code was required for the match. The first approach allowed the identification of 361,242, 16,573, and 279,839 records with current country, historical country, and subregion information, respectively. The second method was the use of a heuristic based on common address field structures to identify “location strings” toward the end of address fields that likely represent countries, then the use of the Python pycountry module to confirm the presence of country information. This approach led to 329,025 records with country information. The third approach was to parse first author email addresses (90,799 records), recover top-level domain information, and use country code Top Level Domain (ccTLD) data from the ISO 3166 Wikipedia page to define countries (72,640 records). Only a subset of email addresses contains country information because some are from companies (.com), nonprofit organizations (.org), and others. Because a large number of records with address fields still did not have country information after taking the above 3 approaches, another approach was implemented to query address fields against a locally installed Nominatim server (v.4.2.3, https://github.com/mediagis/nominatim-docker ) using OpenStreetMap data from GEOFABRIK ( https://www.geofabrik.de/ ) to find locations. Initial testing indicated that the use of full address strings led to false positives, and the computing resource requirement for running the server was high. Thus, only location strings from the second approach that did not lead to country information were used as queries. Because multiple potential matches were returned for each query, the results were sorted based on their location importance values. The above steps led to an additional 72,401 records with country information.
Examining the overlap in country information between approaches revealed that brute force current country and pycountry searches were consistent 97.1% of the time. In addition, both approaches had high consistency with the email-based approach (92.4% and 93.9%). However, brute force subregion and Nominatim-based predictions had the lowest consistencies with the above 3 approaches (39.8% to 47.9%) and each other. Thus, a record’s country information was finalized if the information was consistent between any 2 approaches, except between the brute force subregion and Nominatim searches. This led to 330,328 records with country information.
Topical and country impact metrics
To determine annual country impact, impact scores were determined in the same way as that for annual topical impact, except that values for different countries were calculated instead of topics ( S8 Data ).
Topical preferences by country
To determine topical preference for a country C , a 2 × 2 table was established with the number of records in topic T from C , the number of records in T but not from C , the number of non- T records from C , and the number of non- T records not from C . A Fisher’s exact test was performed for each T and C combination, and the resulting p -values were corrected for multiple testing with the Bejamini–Hochberg method (see S12 Data ). The preference of T in C was defined as the degree of enrichment calculated as log likelihood ratio of values in the 2 × 2 table. Topic 5 was excluded because >50% of the countries did not have records for this topic.
The top 10 countries could be classified into a China–India cluster, an Italy–Spain cluster, and remaining countries (yellow rectangles, Fig 5E ). The clustering of Italy and Spain is partly due to similar research focusing on allergens (topic 0) and mycotoxins (topic 54) and less emphasis on gene family (topic 23) and stress tolerance (topic 28) studies ( Figs 5F and S9 ). There are also substantial differences in topical focus between countries. For example, plant science records from China tend to be enriched in hyperspectral imaging and modeling (topic 9), gene family studies (topic 23), stress biology (topic 28), and research on new plant compounds associated with herbal medicine (topic 69), but less emphasis on population genetics and evolution (topic 86, Fig 5F ). In the US, there is a strong focus on insect pest resistance (topic 75), climate, community, and diversity (topic 83), and population genetics and evolution but less focus on new plant compounds. In summary, in addition to revealing how plant science research has evolved over time, topic modeling provides additional insights into differences in research foci among different countries.
Supporting information
S1 fig. plant science record classification model performance..
(A–C) Distributions of prediction probabilities (y_prob) of (A) positive instances (plant science records), (B) negative instances (non-plant science records), and (C) positive instances with the Medical Subject Heading “Plants” (ID = D010944). The data are color coded in blue and orange if they are correctly and incorrectly predicted, respectively. The lower subfigures contain log10-transformed x axes for the same distributions as the top subfigure for better visualization of incorrect predictions. (D) Prediction probability distribution for candidate plant science records. Prediction probabilities plotted here are available in S13 Data .
https://doi.org/10.1371/journal.pbio.3002612.s001
S2 Fig. Relationships between outlier clusters and the 90 topics.
(A) Heatmap demonstrating that some outlier clusters tend to have high prediction scores for multiple topics. Each cell shows the average prediction score of a topic for records in an outlier cluster. (B) Size of outlier clusters.
https://doi.org/10.1371/journal.pbio.3002612.s002
S3 Fig. Cosine similarities between topics.
(A) Heatmap showing cosine similarities between topic pairs. Top-left: hierarchical clustering of the cosine similarity matrix using the Ward algorithm. The branches are colored to indicate groups of related topics. (B) Topic labels and names. The topic ordering was based on hierarchical clustering of topics. Colored rectangles: neighboring topics with >0.5 cosine similarities.
https://doi.org/10.1371/journal.pbio.3002612.s003
S4 Fig. Relative topical diversity for 20 journals.
The 20 journals with the most plant science records are shown. The journal names were taken from the journal list in PubMed ( https://www.nlm.nih.gov/bsd/serfile_addedinfo.html ).
https://doi.org/10.1371/journal.pbio.3002612.s004
S5 Fig. Topical frequency and top terms during different time periods.
(A-D) Different patterns of topical frequency distributions for example topics (A) 48, (B) 35, (C) 27, and (D) 42. For each topic, the top graph shows the frequency of topical records in each time bin, which are the same as those in Fig 3 (green line), and the end date for each bin is indicated. The heatmap below each line plot depicts whether a term is among the top terms in a time bin (yellow) or not (blue). Blue dotted lines delineate different decades (see S5 Data for the original frequencies, S6 Data for the LOWESS fitted frequencies and the top terms for different topics/time bins).
https://doi.org/10.1371/journal.pbio.3002612.s005
S6 Fig. Prevalence of records mentioning different taxonomic groups in Viridiplantae.
(A, B) Percentage of records mentioning specific taxa at the ( A) major lineage and (B) family levels. (C, D) The prevalence of taxon mentions over time at the (C) major lineage and (E) family levels. The data used for plotting are available in S9 Data .
https://doi.org/10.1371/journal.pbio.3002612.s006
S7 Fig. Changes over time.
(A) Number of genera being mentioned in plant science records during different time bins (the date indicates the end date of that bin, exclusive). (B) Numbers of genera (blue) and organisms (salmon) with draft genomes available from National Center of Biotechnology Information in different years. (C) Percentage of US National Science Foundation (NSF) grants mentioning the genus Arabidopsis over time with peak percentage and year indicated. The data for (A–C) are in S9 Data . (D) Number of plant science records in the top 17 plant science journals from the USA (red), Great Britain (GBR) (orange), India (IND) (light green), and China (CHN) (dark green) normalized against the total numbers of publications of each country over time in these 17 journals. The data used for plotting can be found in S11 Data .
https://doi.org/10.1371/journal.pbio.3002612.s007
S8 Fig. Change in country impact on plant science over time.
(A, B) Difference in 2 impact metrics from 1999 to 2020 for the 10 countries with the highest number of plant science records. (A) H-index. (B) SCImago Journal Rank (SJR). (C, D) Plots show the relationships between the impact metrics (H-index in (C) , SJR in (D) ) averaged from 1999 to 2020 and the slopes of linear fits with years as the predictive variable and impact metric as the response variable for different countries (A3 country codes shown). The countries with >400 records and with <10% missing impact values are included. The data used for plotting can be found in S11 Data .
https://doi.org/10.1371/journal.pbio.3002612.s008
S9 Fig. Country topical preference.
Enrichment scores (LLR, log likelihood ratio) of topics for each of the top 10 countries. Red: overrepresentation, blue: underrepresentation. The data for plotting can be found in S12 Data .
https://doi.org/10.1371/journal.pbio.3002612.s009
S1 Data. Summary of source journals for plant science records, prediction models, and top Tf-Idf features.
Sheet–Candidate plant sci record j counts: Number of records from each journal in the candidate plant science corpus (before classification). Sheet—Plant sci record j count: Number of records from each journal in the plant science corpus (after classification). Sheet–Model summary: Model type, text used (txt_flag), and model parameters used. Sheet—Model performance: Performance of different model and parameter combinations on the validation data set. Sheet–Tf-Idf features: The average SHAP values of Tf-Idf (Term frequency-Inverse document frequency) features associated with different terms. Sheet–PubMed number per year: The data for PubMed records in Fig 1A . Sheet–Plant sci record num per yr: The data for the plant science records in Fig 1A .
https://doi.org/10.1371/journal.pbio.3002612.s010
S2 Data. Numbers of records in topics identified from preliminary topic models.
Sheet–Topics generated with a model based on BioBERT embeddings. Sheet–Topics generated with a model based on distilBERT embeddings. Sheet–Topics generated with a model based on SciBERT embeddings.
https://doi.org/10.1371/journal.pbio.3002612.s011
S3 Data. Final topic model labels and top terms for topics.
Sheet–Topic label: The topic index and top 10 terms with the highest cTf-Idf values. Sheets– 0 to 89: The top 50 terms and their c-Tf-Idf values for topics 0 to 89.
https://doi.org/10.1371/journal.pbio.3002612.s012
S4 Data. UMAP representations of different topics.
For a topic T , records in the UMAP graph are colored red and records not in T are colored gray.
https://doi.org/10.1371/journal.pbio.3002612.s013
S5 Data. Temporal relationships between published documents projected onto 2D space.
The 2D embedding generated with UMAP was used to plot document relationships for each year. The plots from 1975 to 2020 were compiled into an animation.
https://doi.org/10.1371/journal.pbio.3002612.s014
S6 Data. Timestamps and dates for dynamic topic modeling.
Sheet–bin_timestamp: Columns are: (1) order index; (2) bin_idx–relative positions of bin labels; (3) bin_timestamp–UNIX time in seconds; and (4) bin_date–month/day/year. Sheet–Topic frequency per timestamp: The number of documents in each time bin for each topic. Sheets–LOWESS fit 0.1/0.2/0.3: Topic frequency per timestamp fitted with the fraction parameter of 0.1, 0.2, or 0.3. Sheet—Topic top terms: The top 5 terms for each topic in each time bin.
https://doi.org/10.1371/journal.pbio.3002612.s015
S7 Data. Locally weighted scatterplot smoothing (LOWESS) of topical document frequencies over time.
There are 90 scatter plots, one for each topic, where the x axis is time, and the y axis is the document frequency (blue dots). The LOWESS fit is shown as orange points connected with a green line. The category a topic belongs to and its order in Fig 3 are labeled on the top left corner. The data used for plotting are in S6 Data .
https://doi.org/10.1371/journal.pbio.3002612.s016
S8 Data. The 4 criteria used for sorting topics.
Peak: the time when the LOWESS fit of the frequencies of a topic reaches maximum. 1st_reach_thr: the time when the LOWESS fit first reaches a threshold of 60% maximal frequency (peak value). Trend: upward (1), no change (0), or downward (−1). Stable: whether a topic belongs to the stable category (1) or not (0).
https://doi.org/10.1371/journal.pbio.3002612.s017
S9 Data. Change in taxon record numbers and genome assemblies available over time.
Sheet–Genus: Number of records mentioning a genus during different time periods (in Unix timestamp) for the top 100 genera. Sheet–Genus: Number of records mentioning a family during different time periods (in Unix timestamp) for the top 100 families. Sheet–Genus: Number of records mentioning an order during different time periods (in Unix timestamp) for the top 20 orders. Sheet–Species levels: Number of records mentioning 12 selected taxonomic levels higher than the order level during different time periods (in Unix timestamp). Sheet–Genome assembly: Plant genome assemblies available from NCBI as of October 28, 2022. Sheet–Arabidopsis NSF: Absolute and normalized numbers of US National Science Foundation funded proposals mentioning Arabidopsis in proposal titles and/or abstracts.
https://doi.org/10.1371/journal.pbio.3002612.s018
S10 Data. Taxon topical preference.
Sheet– 5 genera LLR: The log likelihood ratio of each topic in each of the top 5 genera with the highest numbers of plant science records. Sheets– 5 genera: For each genus, the columns are: (1) topic; (2) the Fisher’s exact test p -value (Pvalue); (3–6) numbers of records in topic T and in genus X (n_inT_inX), in T but not in X (n_inT_niX), not in T but in X (n_niT_inX), and not in T and X (n_niT_niX) that were used to construct 2 × 2 tables for the tests; and (7) the log likelihood ratio generated with the 2 × 2 tables. Sheet–corrected p -value: The 4 values for generating LLRs were used to conduct Fisher’s exact test. The p -values obtained for each country were corrected for multiple testing.
https://doi.org/10.1371/journal.pbio.3002612.s019
S11 Data. Impact metrics of countries in different years.
Sheet–country_top25_year_count: number of total publications and publications per year from the top 25 countries with the most plant science records. Sheet—country_top25_year_top17j: number of total publications and publications per year from the top 25 countries with the highest numbers of plant science records in the 17 plant science journals used as positive examples. Sheet–prank: Journal percentile rank scores for countries (3-letter country codes following https://www.iban.com/country-codes ) in different years from 1999 to 2020. Sheet–sjr: Scimago Journal rank scores. Sheet–hidx: H-Index scores. Sheet–cite: Citation scores.
https://doi.org/10.1371/journal.pbio.3002612.s020
S12 Data. Topical enrichment for the top 10 countries with the highest numbers of plant science publications.
Sheet—Log likelihood ratio: For each country C and topic T, it is defined as log((a/b)/(c/d)) where a is the number of papers from C in T, b is the number from C but not in T, c is the number not from C but in T, d is the number not from C and not in T. Sheet: corrected p -value: The 4 values, a, b, c, and d, were used to conduct Fisher’s exact test. The p -values obtained for each country were corrected for multiple testing.
https://doi.org/10.1371/journal.pbio.3002612.s021
S13 Data. Text classification prediction probabilities.
This compressed file contains the PubMed ID (PMID) and the prediction probabilities (y_pred) of testing data with both positive and negative examples (pred_prob_testing), plant science candidate records with the MeSH term “Plants” (pred_prob_candidates_with_mesh), and all plant science candidate records (pred_prob_candidates_all). The prediction probability was generated using the Word2Vec text classification models for distinguishing positive (plant science) and negative (non-plant science) records.
https://doi.org/10.1371/journal.pbio.3002612.s022
Acknowledgments
We thank Maarten Grootendorst for discussions on topic modeling. We also thank Stacey Harmer, Eva Farre, Ning Jiang, and Robert Last for discussion on their respective research fields and input on how to improve this study and Rudiger Simon for the suggestion to examine differences between countries. We also thank Mae Milton, Christina King, Edmond Anderson, Jingyao Tang, Brianna Brown, Kenia Segura Abá, Eleanor Siler, Thilanka Ranaweera, Huan Chen, Rajneesh Singhal, Paulo Izquierdo, Jyothi Kumar, Daniel Shiu, Elliott Shiu, and Wiggler Catt for their good ideas, personal and professional support, collegiality, fun at parties, as well as the trouble they have caused, which helped us improve as researchers, teachers, mentors, and parents.
- View Article
- PubMed/NCBI
- Google Scholar
- 2. Blei DM, Lafferty JD. Topic Models. In: Srivastava A, Sahami M, editors. Text Mining. Cambridge: Chapman and Hall/CRC; 2009. pp. 71–93.
- 7. ChatGPT. [cited 2023 Aug 25]. Available from: https://chat.openai.com
- 9. Fei-Fei L, Perona P. A Bayesian hierarchical model for learning natural scene categories. 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05); 2005. pp. 524–531 vol. 2. https://doi.org/10.1109/CVPR.2005.16
- 19. Blei DM, Lafferty JD. Dynamic topic models. Proceedings of the 23rd International Conference on Machine learning. New York, NY, USA: Association for Computing Machinery; 2006. pp. 113–120. https://doi.org/10.1145/1143844.1143859
- 35. Kuhn T. The Structure of Scientific Revolution. Chicago: University of Chicago Press; 1962.
- 36. CiteSeer | Proceedings of the second international conference on Autonomous agents. [cited 2023 Aug 23]. Available from: https://dl.acm.org/doi/10.1145/280765.280786
- 39. Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York, NY, USA: ACM; 2016. pp. 785–794. https://doi.org/10.1145/2939672.2939785
- 40. Řehůřek R, Sojka P. Software Framework for Topic Modelling with Large Corpora. Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks. Valletta, Malta: ELRA; 2010. pp. 45–50.
- 42. Hugging Face–The AI community building the future. 2023 Aug 19 [cited 2023 Aug 25]. Available from: https://huggingface.co/

Growing Beyond Earth Annual Student Research Symposium
NASA Science Activation's Growing Beyond Earth project is a 6th-12th grade classroom-based citizen science project developed by Fairchild Tropical Botanic Garden in partnership with scientists at NASA, designed to advance NASA research on growing plants in space. It includes a series of plant experiments conducted by students in a Fairchild-designed plant habitat similar to the Vegetable Production System (Veggie) on the International Space Station. Tens of thousands of middle and high school students and their teachers nationwide have contributed hundreds of thousands of data points and tested 180 varieties of edible plants for NASA, with four having been flown on the International Space Station based on the student's results.
On April 20, 2024, 400+ students and teachers from 75 schools across the nation (and world) gathered at Fairchild Tropical Botanic Garden to participate in the annual Growing Beyond Earth Student Research Symposium. Participants presented their original ISS-analogue space crop research in-person to a panel of judges from NASA Kennedy Space Center and/or virtually to two dozen university and NASA scientists. Experiments ranged from “Effect of Sargassum Extract Fertilizer on Growth of Chervil Vertissimo” to “Magnetoelectric Effect on Purple Magic Pac Choi”. A big congratulations to all the teacher and student scientists who presented their research and much heartfelt appreciation to all involved in Growing Beyond Earth for inspiring and encouraging the next generation of explorers.
Middle or high school teachers interested in bringing Growing Beyond Earth Citizen Science into your classrooms are invited to join the upcoming information sessions, during which the Growing Beyond Earth team will explain the program and its implementation in the classroom (followed by a Q&A session).
Two Dates Available: June 10, 7-8 p.m. EST: Join the Webinar July 1, 11 a.m. - 12 p.m. EST: Join the Webinar
The Growing Beyond Earth project is supported by NASA under cooperative agreement award number NNH21ZDA001N-SciAct and is part of NASA’s Science Activation Portfolio. Learn more about how Science Activation connects NASA science experts, real content, and experiences with community leaders to do science in ways that activate minds and promote deeper understanding of our world and beyond: https://science.nasa.gov/learn

Related Terms
- Biological & Physical Sciences
- Citizen Science
- Grades 5 - 8 for Educators
- Grades 9-12 for Educators
- Opportunities For Educators to Get Involved
- Opportunities For Students to Get Involved
- Science Activation
Explore More

Space Station Research Advances NASA’s Plans to Explore the Moon, Mars
Space, the saying goes, is hard. And the farther humans go, the harder it can get. Some of the challenges on missions to explore the Moon and Mars include preventing microbial contamination of these destinations, navigating there safely, protecting crew members and hardware from radiation, and maintaining and repairing equipment. Research on the International Space […]

The Moon and Amaey Shah

Arizona Students Go on an Exoplanet Watch
Discover more topics from nasa.
James Webb Space Telescope

Perseverance Rover

Parker Solar Probe


Research Topics
Plant and insect biodiversity can contribute to crucial ecosystem services in agricultural landscapes, including pollination and biological control.
The goal of our research is to inform the development of integrated, multi-tactical weed and crop management strategies that are both economically and ecologically sustainable.

Our lab’s interest in weedy plants in agroecosystems is paralleled by our work on invasive species in forest systems. We are studying population dynamics and spatial distributions of forest invasive species including Microstegium vimieum (Japanese stiltgrass).

We are interested in the pattern and process of dispersal and how dispersal impacts management of invasive weed species. Currently several projects in the lab examine seed transported along roads and in the atmosphere.

- Degrees & Programs
- College Directory
Information for
- Faculty & Staff
- Visitors & Public
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
Plant genetics articles from across Nature Portfolio
Plant genetics deals with heredity in plants, specifically mechanisms of hereditary transmission and variation of inherited characteristics. Plant genetics differs from animal genetics in a number of ways: somatic mutations can contribute to the germ line more easily as flowers develop at the end of branches composed of somatic cells; polyploidy is more common; and plants additionally contain chloroplastic DNA.
Raison d’être of the plastid repeat
Creation of a plastid genome without an inverted repeat in Nicotiana tabacum reveals a role for the inverted repeat in gene dosage and the regulation of replication by total DNA content rather than copy number.
- Jeffrey P. Mower
How cauliflower got its curd
For a variety of reasons, genetic understanding of the steps leading to domestication of the nutrient-rich edible arrested inflorescence of cauliflower — its curd — has proven relatively intractable. A genomic study now unravels the details.
- Alisdair R. Fernie
- Mustafa Bulut
Clonal gametes enable polyploid genome design
Many plant products eaten daily in human diets — such as potato or banana — are polyploid and are notoriously difficult to breed. In this study, the fusion of clonal gametes from distinct diploid tomato parents is used as a blueprint for the design of polyploid genomes in crops.
Related Subjects
- Genome duplication
- Plant hybridization
- Polyploidy in plants
- Transgenic plants
Latest Research and Reviews
Targeted suppression of siRNA biogenesis in Arabidopsis pollen promotes triploid seed viability
In Arabidopsis, the pollen vegetative cell is regarded as a source of mobile siRNAs that guide male germline reprogramming. This study demonstrates that siRNA triggers of triploid seed lethality originate in germline companion cells after meiosis.
- Kannan Pachamuthu
- Matthieu Simon
- Filipe Borges
Genome resources for three modern cotton lines guide future breeding efforts
Analyses of three newly sequenced modern cultivar cotton genomes revealed sequence and structural variation alongside traces of ancient and ongoing introgressions. Moreover, transcriptome analysis pointed at unique fibre quality traits of cultivars.
- Avinash Sreedasyam
- John T. Lovell
- Jeremy Schmutz
Removal of the large inverted repeat from the plastid genome reveals gene dosage effects and leads to increased genome copy number
Removing the large inverted repeat region from the chloroplast genome revealed a gene dosage benefit for the ribosomal RNA operon. The reduced genome size resulted in increased genome copy numbers and offers potential for synthetic biology.
- Carolin Krämer
- Christian R. Boehm
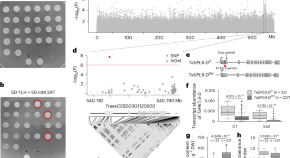
Variation in TaSPL6-D confers salinity tolerance in bread wheat by activating TaHKT1;5-D while preserving yield-related traits
The study identifies TaSPL6-D as a negative regulator of TaHKT1;5-D and salinity tolerance in bread wheat. An insertion variation of TaSPL6-D , mainly hidden in landraces, shows the potential for breeding salt-tolerant crops.
- Weiming Shi
Seedling root system adaptation to water availability during maize domestication and global expansion
Characterization of seminal root number variation in the root systems of >9,000 global maize accessions and its wild relatives provides insights into root trait adaptation to environments during domestication and global expansion.
- Frank Hochholdinger
Circadian and photoperiodic regulation of the vegetative to reproductive transition in plants
A review synthesizes interplays between photoperiodism and circadian regulation of the vegetative to reproductive transition in Arabidopsis and crops in responses to changing day lengths that ensures optimal timing for flowering and seed production.
- Tongwen Han
- Z. Jeffrey Chen
News and Comment

Coffee history under the genomic lens
Polyploid sugarcane genome architecture.

A sweet victory for sugarcane genomics
Owing to its size and complexity, the genome of modern sugarcane has never been previously assembled in its entirety, which leaves it as one of the last remaining major crop species without a reference genome. The newly completed polyploid assembly of an archetypal modern hybrid reveals the complexities of sugarcane’s genetic past, and presents new opportunities for the researchers and breeders invested in its future.
- Elizabeth A. Cooper
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
- Made in America
Part 1: Made in America research shows enhanced production efficiencies
Enhanced production efficiency and growth opportunities are among perceived benefits of a “made in america strategy," noted in a spring 2024 “made in america and reshoring report” from control engineering and plant engineering..

Learning Objectives
- Understand how “Made in America” research from WTWH Media (including Control Engineering, Plant Engineering ) was conducted and how subscribers define it and their understanding of its importance.
- Learn about made in America opportunities, such as U.S. manufacturing expansion and reshoring.
- Explore made in America challenges, including competition, higher production costs, labor challenges and production efficiency, topics that can be helped by automation. A part-2 “Made in America” research article expands with information on manufacturing supply chain lead times, innovation, strengths and weaknesses, quality and costs.
Part 1: Made in America manufacturing challenges, opportunities insights
- Made in America research from WTWH Media (including Control Engineering, Plant Engineering ) asked subscribers responding to the survey to define “Made in America” and explain its importance.
- Made in America opportunities include U.S. manufacturing expansion and reshoring.
- Made in America challenges include competition, higher production costs, labor challenges and production efficiency, topics that can be helped by automation. A part-2 “Made in America” research article expands with information on manufacturing supply chain lead times, innovation, strengths and weaknesses, quality and costs.
Made in America and reshoring efforts are perceived positively for manufacturing competitiveness and growth by respondents to WTWH Media research. For the “Made in America and Reshoring Report,” Control Engineering and Plant Engineering publications conducted original research to study the impact of made in America and reshoring efforts on manufacturing competitiveness and resiliency topics. Among key topics covered in part 1 of this article are made in America definition, how it’s important, areas of growth, critical challenges and how made in American influences manufacturing production efficiency.
Made in America enhances production efficiency according to 61% of respondents; of that, 34% said it moderately enhances efficiency, and 17% said it significantly enhances efficiency. No significant change was observed by 32%; 10% said not applicable, and 7% sees a reduction in production efficiency as a result of made in America.
Methodology for the made in America and reshoring survey
Research for the 2024 Made in America and Reshoring Report resulted from an emailed survey to subscribers, producing 320 qualified responses during spring 2024, for a margin of error of +/- 5.5% at a 95% confidence level.
Made in America: What is it and why is it important?
Because made and assembled differ in meanings, the research asked subscribers to explain their understanding of what is “made in America” (Figure 1). A majority (53%) said all parts are made and assembled in the USA, while 21% said it’s OK for a majority of parts to be made and assembled in the USA and 14% more said a majority of parts, including the defining part, and assembled in the USA. Six other combinations accounted for 5% or less each.
Figure 1 asked subscribers how they define “Made in America” including where parts are sourced and where products are assembled. Some respondents think majority, rather than all, parts sources in USA would be OK, in the spring 2024 “Made in America and Reshoring Report,” from Control Engineering and Plant Engineering. Courtesy: WTWH Media research
The made in America label (Figure 2) is absolutely critical (12%) or very important (42%) to more than half (54%) of those responding. In addition, 30% more called it moderately important.
Figure 2: More than 70% of respondents said the “Made in America” label is important in decision-making processes for sourcing products and services, in the spring 2024 “Made in America and Reshoring Report,” from Control Engineering and Plant Engineering. Courtesy: WTWH Media research
Made in America: Opportunities for U.S. manufacturing expansion, reshoring
A diverse range of sectors (Figure 3) have “the greatest growth opportunities” due to made in America initiatives. Top five areas, from 50% to 46%, were computers, electronics and communications; aerospace; machinery; electrical equipment; and automotive and transportation. Respondents identified half of 24 sectors greater than 30%. None ranked lower than 20%.
Figure 3: Some sectors are perceived as having greater growth opportunities from “Made in America” initiatives, including computer and communications, aerospace, machinery, electrical equipment and automotive, in the spring 2024 “Made in America and Reshoring Report,” from Control Engineering and Plant Engineering. Courtesy: WTWH Media research
The “made in America” theme influences views on reshoring (Figure 4) in a very positive way for 42% of those responding, somewhat positively for 33% more; 18% said they were neutral; 5% said it was complex, depending on center. Only 2% viewed it negatively.
Figure 4: The “Made in America” theme has a positive influence on reshoring for three-quarters of survey respondents, in the spring 2024 “Made in America and Reshoring Report,” from Control Engineering and Plant Engineering. Courtesy: WTWH Media research
Made in America challenges: Competition, higher production costs, labor
When adopting a “made in America” strategy, one challenge is significantly greater, and that is competition with low-cost international markets at 80% (Figure 5). Among other challenges are elevated production costs at 59%, scarcity of skilled labor at 50%, regulatory compliance difficulties at 47% and limited supply chain resources at 46%. Lower on the list was volatility in consumer demand at 21% and scalability of operations at 20%. Automation can help with five of seven of the challenges.
Figure 5: The largest challenge by far when adopting a “Made in America” strategy is competition with low-cost international markets, followed by elevated production costs, in the spring 2024 “Made in America and Reshoring Report,” from Control Engineering and Plant Engineering. Courtesy: WTWH Media research
Made in America impact on production efficiency, research article part 2 topics
“Made in America” significantly (17%) or moderately enhances production efficiency (34%) for more than half (51%) of those responding. Among other replies (Figure 6), 32% perceived no significant change, 10% didn’t think it was applicable, and 7% said made in America moderately (5%) or significantly (2%) reduces efficiency.
Figure 6: More than half of respondents said “Made in America” enhanced production efficiency in their facilities, in the spring 2024 “Made in America and Reshoring Report,” from Control Engineering and Plant Engineering. Courtesy: WTWH Media research
Part 2 article on this research will answer questions about supply chain resilience, lead times, innovation, strengths, weaknesses, quality and quality versus cost.
You asked; we responded with more “made in America” content
Because more than half of respondents said they like to learn more about topics related to made in America, Control Engineering, Plant Engineering and Consulting-Specifying Engineer magazines made an area on their websites for related “Made in America” content.
Mark T. Hoske is editor-in-chief, Control Engineering, WTWH Media , [email protected] . Amanda McLeman , marketing research manager, WTWH Media, conducted the research.
KEYWORDS: Made in America, manufacturing automation, manufacturing efficiency
CONSIDER THIS
How does automation help with made in America, quality and efficiency?
Original content can be found at Control Engineering .
Do you have experience and expertise with the topics mentioned in this content? You should consider contributing to our CFE Media editorial team and getting the recognition you and your company deserve. Click here to start this process.
Privacy Overview
Main Content
Plant science - research topics.
The following Research Topics are led by experts in their field and contribute to the scientific understanding of plant-science. These Research topics are published in the peer-reviewed journal Frontiers in Plant Science , as open access articles .

Utilizing Machine Learning with Phenotypic and Genotypic Data to enhance Effective Breeding in Agricultural and Horticultural Crops
Changing climate is driving frequent extreme weather events such as increased drought, and unpredictable seasonal precipitation, resulting in increased salinity, and shifts in pest pressures. Furthermore, changing climate has resulted in shifts in gr...

The Regulation of Secondary Plant Cell Wall Formation
Secondary cell walls (SCWs) are the main constituent of plant biomass. SCWs are synthesized in specific cells such as tracheary elements and fibers. These SCWs not only provide mechanical support to the plant body and enable long-distance water trans...

Plant Parasitic Nematode–Host Interactions: Mechanisms and Exploitative Management Strategies
Plant parasitic nematodes (PPN) are pathogens common in global agricultural systems. There is at least one species of PPN for all major food crops and yield losses caused by nematodes threaten global food security. Management of PPN is challenging an...

Virome Analysis for the Identification of Emerging and Re-Emerging Viruses in Plants
The advent of next-generation sequencing (NGS) technology, including pyrosequencing (454; Roche) and sequencing by synthesis (Solexa genome analyzer; Illumina), has facilitated the use of metagenomic analyses to characterize viruses in many host plan...

Chilling Tolerance and Regulation of Horticultural Crops: Physiological, Molecular, and Genetic Perspectives
Chilling injury affects crops in the tropical and subtropical regions. Damage can include surface pitting, discolouration, internal breakdown, water soaking, failure to ripen, growth inhibition, wilting, loss of flavour, and decay. Post-harvest handl...

Applicative and Ecological Aspects of Mycorrhizal Symbioses
Mycorrhizal symbioses, a mutual association between plants and fungi plays a vital role in shaping and balancing the ecosystem of our planet. Approximately 80% of vascular plant species form mycorrhizal associations between their roots and soil borne...

Recent Advances in Research and Development for Vegetable Crops Under Protected Cultivation
Protected cropping shelters crops from extreme climatic conditions by modifying the internal growing conditions in their favor. The technology has widely been used for vegetable crops in different climates such as temperate, tropical, subtropical, ar...

Infection and Colonization of Horticultural Crops by Microbial Pathogens
Horticultural crops are an integral part of economic growth and they contribute significantly to agricultural production. Plants can be infected by pathogenic oomycetes, fungi and bacteria that cause significant yield losses worldwide and have a grea...

Current Advances in Botrytis cinerea: Biology, Pathogenesis and Interaction with Host Plants
Better known to wine connoisseurs as the “noble rot”, Botrytis cinerea is indeed an important plant pathogen with a very diverse host range, causing gray mold disease in over 1000 plant species, including most vegetable and fruit crops, trees, and fl...

COMMENTS
Analyses of three newly sequenced modern cultivar cotton genomes revealed sequence and structural variation alongside traces of ancient and ongoing introgressions. Moreover, transcriptome analysis ...
By Rachel Berkowitz April 9, 2024. Animals. Big monarch caterpillars don't avoid toxic milkweed goo. They binge on it. Instead of nipping milkweed to drain the plants' defensive sap, older ...
Response and Adaptation of Terrestrial Ecosystem Carbon, Nitrogen, and Water Cycles to Climate Change in Arid Desert Regions. Jianping Li. Huang Lei. Kaibo Wang. Dafeng Hui. 1,012 views. The most cited plant science journal advances our understanding of plant biology for sustainable food security, functional ecosystems and human health.
Summary The 'One Hundred Important Questions Facing Plant Science Research' project aimed to capture a global snapshot of the current issues and future questions facing plant science. ... there was remarkable consensus on the most important topics. The clearest outcome of the 100 Plant Science Questions project in 2022 was the prominence of ...
The importance of early human choices of wild plants in determining crop physiology. Leaf ecophysiological traits of crops are primarily inherited from their wild progenitors, challenging the ...
Abiotic stresses challenge plant growth. In response, plants often rapidly accumulate proline. This study reveals the filament structures of plant P5CS, the key enzyme in proline synthesis ...
This project, led by Prof. Claire Grierson (University of Bristol, UK),is a novel piece of research that gathered over 600 questions about plant science, from botanically curious members of the public to scientific and industrial leaders around the world. A team of 20 plant scientists from 15 nations were assembled to identify 100 of the most ...
Many plants have developed physical and chemical defenses to fend off herbivores. A well-known strategy in flowering plants is to produce nectar to attract "ant bodyguards." Recent research ...
This editorial initiative of particular relevance, led by Dr. Anna Stepanova, Specialty Chief Editor of the Plant Physiology section, is focused on new insights, novel developments, current challenges, latest discoveries, recent advances, and future perspectives in the field of plant physiology.<br/><br/>The Research Topic solicits brief ...
100+ Botany Research Topics [Updated 2024] General / By StatAnalytica / 5th January 2024. Botany, the scientific study of plants, holds the key to understanding the intricate and fascinating world of flora that surrounds us. As we delve into the realm of botany research, we uncover a vast array of botany research topics that not only contribute ...
The Plant Biotechnology section at Frontiers in Plant Science mainly publishes applied studies examining how plants can be improved using modern genetic techniques (Lloyd and Kossmann, 2021). This Research Topic was designed to allow editors from the section to highlight some of their own plant biotechnological work.
The Plant Cell publishes novel research of special significance in plant biology, especially in the areas of cellular biology, molecular biology, biochemistry, genetics, development, and evolution.
Before the panel meeting the full list of 350 submitted questions was roughly organized into groups according to topic. Each panel member independently selected their top 20 questions and these lists were combined. ... the panel decided to categorize the questions into five broad areas that reflect the breadth and depth of plant research ...
Explore our most highly accessed plant science articles in 2017. Featuring authors from around the World, these papers highlight valuable research within plant science from an international community.
23 Ideas for Science Experiments Using Plants. Plants are tremendously crucial to life on earth. They are the foundation of food chains in almost every ecosystem. Plants also play a significant role in the environment by influencing climate and producing life-giving oxygen. Plant project studies allow us to learn about plant biology and ...
The Department of Plant Science conducts critical research targeting plant biology, ecology, genetics, cultivation, human uses, sustainability, and more. We are proud to be part of a Tier 1 Research University, devoting the resources needed to maintain world-class facilities and attract talented faculty, staff, and students.
Credit: Pixabay/CC0 Public Domain. Irish researchers involved in cataloging the world's plant species are hunting for answers as to what makes some groups so successful. One of their major goals ...
These topics represent long-standing plant science research foci, including studies of plant physiology (topics 4, 58, and 81), genetics (topic 61), and medicinal plants (topic 53). The second category contains 8 topics established before the 1980s that have mostly decreased in prevalence since (the early category, Fig 3B). Two examples are ...
Science Fair Project Idea. Plants move—not very quickly compared to animals, but they do move. Their roots grow downward in response to gravity, and their stems grow upward toward the Sun. In this plant biology science fair project, you will investigate how young plants respond through movement to light. Read more.
Plant biology is a key area of science that bears major weight in the mankind's ongoing and future efforts to combat the consequences of global warming, climate change, pollution, and population growth. An in-depth understanding of plant physiology is paramount to our ability to optimize current agricultural practices, to develop new crop ...
Plant Biology Topics. Plant biology research in our lab can be broken down into: Plant Development. Regulation of Gene Expression. Mechanisms of Plant Defense Responses. Key Metabolic Pathways. Genetic Transformation. Starch Biosynthesis.
Haploids fast-track hybrid plant breeding. Two studies report the use of paternal haploids to enable one-step transfer of cytoplasmic male sterility in maize and broccoli, which resolves a key ...
NASA Science Activation's Growing Beyond Earth project is a 6th-12th grade classroom-based citizen science project developed by Fairchild Tropical Botanic Garden in partnership with scientists at NASA, designed to advance NASA research on growing plants in space. It includes a series of plant experiments conducted by students in a Fairchild-designed plant habitat similar to the Vegetable ...
Invasive Species Research. Our lab's interest in weedy plants in agroecosystems is paralleled by our work on invasive species in forest systems. We are studying population dynamics and spatial distributions of forest invasive species including Microstegium vimieum (Japanese stiltgrass).
Plant ecology is a sub-discipline of ecology focussed on the distribution and abundance of plants, and their interactions with the biotic and abiotic environment. Latest Research and Reviews
Calcium (Ca2+) signaling is involved in the generation of Ca2+ signatures against various stimuli in plants. However, there are certain Ca2+ sensors that sense and decode that signature in downstream Ca2+ signaling, and those Ca2+ responder proteins include calmodulins, calmodulin-like proteins, calcium-dependent protein kinases, and calcineurin B-like proteins Calmodulins (CaMs). On the other ...
RSS Feed. Plant genetics deals with heredity in plants, specifically mechanisms of hereditary transmission and variation of inherited characteristics. Plant genetics differs from animal genetics ...
Made in America impact on production efficiency, research article part 2 topics. "Made in America" significantly (17%) or moderately enhances production efficiency (34%) for more than half (51%) of those responding. Among other replies (Figure 6), 32% perceived no significant change, 10% didn't think it was applicable, and 7% said made in ...
The following Research Topics are led by experts in their field and contribute to the scientific understanding of plant-science. These Research topics are published in the peer-reviewed journal Frontiers in Plant Science, as open access articles.