- How It Works
- PhD thesis writing
- Master thesis writing
- Bachelor thesis writing
- Dissertation writing service
- Dissertation abstract writing
- Thesis proposal writing
- Thesis editing service
- Thesis proofreading service
- Thesis formatting service
- Coursework writing service
- Research paper writing service
- Architecture thesis writing
- Computer science thesis writing
- Engineering thesis writing
- History thesis writing
- MBA thesis writing
- Nursing dissertation writing
- Psychology dissertation writing
- Sociology thesis writing
- Statistics dissertation writing
- Buy dissertation online
- Write my dissertation
- Cheap thesis
- Cheap dissertation
- Custom dissertation
- Dissertation help
- Pay for thesis
- Pay for dissertation
- Senior thesis
- Write my thesis

119 Genetics Research Topics You Must Know About

Put simply, Genetics is the study of genes and hereditary traits in living organisms. Knowledge in this field has gone up over time, and this is proportional to the amount of research.
Right from the DNA structure discovery, a lot more has come out into the open. There are so many genetics research topics to choose from because of the wide scope of research done in recent years.
Genetics is so dear to us since it helps us understand our genes and hereditary traits. In this guide, you will get to understand this subject more and get several topic suggestions that you can consider when looking for interesting genetics topics.
Writing a paper on genetics is quite intriguing nowadays. Remember that because there are so many topics in genetics, choosing the right one is crucial. It will help you cut down on research time and the technicality of selecting content for the topic. Thus, it would matter a lot if you confirmed whether or not the topic you’re choosing has relevant sources in plenty.
What Is Genetics?
Before we even go deeper into genetics topics for research papers, it is essential to have a basic understanding of what the subject entails.
Genetics is a branch of Biology to start with. It is mainly focused on the study of genetic variation, hereditary traits, and genes.
Genetics has relations with several other subjects, including biotechnology, medicine, and agriculture. In Genetics, we study how genes act on the cell and how they’re transmitted from a parent to the offspring. In modern Genetics, the emphasis is more on DNA, which is the chemical substance found in genes. Remember that Genetics cut across animals, insects, and plants – basically any living organism there is.
Tips On How To Write A Decent Research Paper On Genetics
When planning to choose genetics topics, you should also make time and learn how to research. After all, this is the only way you can gather the information that will help you come up with the content for the paper. Here are some tips that can bail you out whenever you feel stuck:
Choosing the topic, nonetheless, is not an easy thing for many students. There are just so many options present, and often, you get spoilt for choice. But note that this is an integral stage/process that you have to complete. Do proper research on the topic and choose the kind of information that you’d like to apply.
Choose a topic that has enough sources academically. Also, choosing interesting topics in genetics is a flex that can help you during the writing process.
On the web, there’s a myriad of information that often can become deceiving. Amateurs try their luck to put together several pieces of information in a bid to try and convince you that they are the authority on the subject. Many students become gullible to such tricks and end up writing poorly in Genetics.
Resist the temptation to look for an easy way of gaining sources/information. You have to take your time and dig up information from credible resources. Otherwise, you’ll look like a clown in front of your professor with laughable Genetics content.
Also, it is quite important that you check when your sources were updated or published. It is preferred and advised that you use recent sources that have gone under satisfactory research and assessment.
Also, add a few words to each on what you’re planning to discuss.Now, here are some of the top genetics paper topics that can provide ideas on what to write about.
Good Ideas For Genetics Topics
Here are some brilliant ideas that you can use as research paper topics in the Genetics field:
- Is the knowledge of Genetics ahead of replication and research?
- What would superman’s genetics be like?
- DNA molecules and 3D printing – How does it work?
- How come people living in mountainous regions can withstand high altitudes?
- How to cross genes in distinct animals.
- Does gene-crossing really help to improve breeds or animals?
- The human body’s biggest intriguing genetic contradictions
- Are we still far away from achieving clones?
- How close are we to fully cloning human beings?
- Can genetics really help scientists to secure various treatments?
- Gene’s regulation – more details on how they can be regulated.
- Genetic engineering and its functioning.
- What are some of the most fascinating facts in the field of Genetics?
- Can you decipher genetic code?
- Cancer vaccines and whether or not they really work.
- Revealing the genetic pathways that control how proteins are made in a bacterial cell.
- How food affects the human body’s response to and connection with certain plants’ and animals’ DNA.
Hot Topics In Genetics
In this list are some of the topics that raise a lot of attention and interest from the masses. Choose the one that you’d be interested in:
- The question of death: Why do men die before women?
- Has human DNA changed since the evolution process?
- How much can DNA really change?
- How much percentage of genes from the father goes to the child?
- Does the mother have a higher percentage of genes transferred to the child?
- Is every person unique in terms of their genes?
- How does genetics make some of us alike?
- Is there a relationship between diets and genetics?
- Does human DNA resemble any other animal’s DNA?
- Sleep and how long you will live on earth: Are they really related?
- Does genetics or a healthy lifestyle dictate how long you’ll live?
- Is genetics the secret to long life on earth?
- How much does genetics affect your life’s quality?
- The question on ageing: Does genetics have a role to play?
- Can one push away certain diseases just by passing a genetic test?
- Is mental illness continuous through genes?
- The relationship between Parkinson’s, Alzheimer’s and the DNA.
Molecular Genetics Topics
Here is a list of topics to help you get a better understanding of Molecular genetics:
- Mutation of genes and constancy.
- What can we learn more about viruses, bacteria, and multicellular organisms?
- A study on molecular genetics: What does it involve?
- The changing of genetics in bacteria.
- What is the elucidation of the chemical nature of a gene?
- Prokaryotes genetics: Why does this take a centre stage in the genetics of microorganisms?
- Cell study: How this complex assessment has progressed.
- What tools can scientists wield in cell study?
- A look into the DNA of viruses.
- What can the COVID-19 virus help us to understand about genetics?
- Examining molecular genetics through chemical properties.
- Examining molecular genetics through physical properties.
- Is there a way you can store genetic information?
- Is there any distinction between molecular levels and subcellular levels?
- Variability and inheritance: What you need to note about living things at the molecular level.
- The research and study on molecular genetics: Key takeaways.
- What scientists can do within the confines of molecular genetics?
- Molecular genetics research and experiments: What you need to know.
- What is molecular genetics, and how can you learn about it?
Human Genetics Research Topics
Human genetics is an interesting field that has in-depth content. Some topics here will jog your brain and invoke curiosity in you. However, if you have difficulty writing a scientific thesis , you can always contact us for help.
- Can you extend your life by up to 100% just by gaining more understanding of the structure of DNA?
- What programming can you do with the help of DNA?
- Production of neurotransmitters and hormones through DNA.
- Is there something that you can change in the human body?
- What is already predetermined in the human body?
- Do genes capture and secure information on someone’s mentality?
- Vaccines and their effect on the DNA.
- What’s the likelihood that a majority of people on earth have similar DNA?
- Breaking of the myostatin gene: What impact does it have on the human body?
- Is obesity passed genetically?
- What are the odds of someone being overweight when the rest of his lineage is obese?
- A better understanding of the relationship between genetics and human metabolism.
- The truths and myths engulfing human metabolism and genetics.
- Genetic tests on sports performance: What you need to know.
- An insight on human genetics.
- Is there any way that you can prevent diseases that are transmitted genetically?
- What are some of the diseases that can be passed from one generation to the next through genetics?
- Genetic tests conducted on a person’s country of origin: Are they really accurate?
- Is it possible to confirm someone’s country of origin just by analyzing their genes?
Current Topics in Genetics
A list to help you choose from all the most relevant topics:
- DNA-altering experiments: How are scientists conducting them?
- How important is it to educate kids about genetics while they’re still in early learning institutions?
- A look into the genetics of men and women: What are the variations?
- Successes and failures in the study of genetics so far.
- What does the future of genetics compare to the current state?
- Are there any TV series or science fiction films that showcase the future of genetics?
- Some of the most famous myths today are about genetics.
- Is there a relationship between genetics and homosexuality?
- Does intelligence pass through generations?
- What impact does genetics hold on human intelligence?
- Do saliva and hair contain any genetic data?
- What impact does genetics have on criminality?
- Is it possible that most criminals inherit the trait through genetics?
- Drug addiction and alcohol use: How close can you relate it to genetics?
- DNA changes in animals, humans, and plants: What is the trigger?
- Can you extend life through medication?
- Are there any available remedies that extend a person’s life genetically?
- Who can study genetics?
- Is genetics only relevant to scientists?
- The current approach to genetics study: How has it changed since ancient times?
Controversial Genetics Topics
Last, but definitely not least, are some controversial topics in genetics. These are topics that have gone through debate and have faced criticism all around. Here are some you can write a research paper about:
- Gene therapy: Some of the ethical issues surrounding it.
- The genetic engineering of animals: What questions have people raised about it?
- The controversy around epigenetics.
- The human evolution process and how it relates to genetics.
- Gene editing and the numerous controversies around it.
- The question on same-sex relations and genetics.
- The use of personal genetic information in tackling forensic cases.
- Gene doping in sports: What you need to know.
- Gene patenting: Is it even possible?
- Should gene testing be compulsory?
- Genetic-based therapies and the cloud of controversy around them.
- The dangers and opportunities that lie in genetic engineering.
- GMOs and their impact on the health and welfare of humans.
- At what stage in the control of human genetics do we stop to be human?
- Food science and GMO.
- The fight against GMOs: Why is it such a hot topic?
- The pros and cons of genetic testing.
- The debates around eugenics and genetics.
- Labelling of foods with GMO: Should it be mandatory?
- What really are the concerns around the use of GMOs?
- The Supreme Court decision on the patent placed on gene discoveries.
- The ethical issues surrounding nurses and genomic healthcare.
- Cloning controversial issues.
- Religion and genetics.
- Behavior learning theories are pegged on genetics.
- Countries’ war on GMOs.
- Studies on genetic disorders.
Get Professional Help Online
Now that we have looked at the best rated topics in genetics, from interesting to controversial topics genetics, you have a clue on what to choose. These titles should serve as an example of what to select.
Nonetheless, if you need help with a thesis, we are available to offer professional and affordable thesis writing services . Our high quality college and university assignment assistance are available to all students online at a cheap rate. Get a sample to check on request and let us give you a hand when you need it most.

Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Comment * Error message
Name * Error message
Email * Error message
Save my name, email, and website in this browser for the next time I comment.
As Putin continues killing civilians, bombing kindergartens, and threatening WWIII, Ukraine fights for the world's peaceful future.
Ukraine Live Updates

- Vision, Mission and Purpose
- Focus & Scope
- Editorial Info
- Open Access Policy
- Editing Services
- Article Outreach
- Why with us
- Focused Topics
- Manuscript Guidelines
- Ethics & Disclosures
- What happens next to your Submission
- Submission Link
- Special Issues
- Latest Articles
- All Articles
- FOCUSED RESEARCH TOPICS
| Twin and family studies |
| Measured genetic variants |
| Quasi-experimental designs |
| Genetic influences on behaviour |
| Nature of environmental influence |
| Nature of genetic influence |
| Psychiatric genetics |
| Karyotyping |
| Banding technique |
| Comparative genome hybridization |
| FISH (fluorescent in situ hybridization) |
| Molecular basis |
| DNA damage |
| Techniques used to study epigenetics |
| ChIP-on-chip and ChIP-Seq) |
| Fluorescent in situ hybridization |
| Methylation-sensitive restriction enzymes |
| DNA adenine methyltransferase identification (DamID) |
| Bisulfite sequencing |
| Mechanisms |
| Covalent modifications |
| RNA transcripts |
| MicroRNAs |
| mRNA |
| sRNAs |
| Prions |
| Structural inheritance |
| Nucleosome positioning |
| Functions and consequences |
| Development |
| Transgenerational |
| Epigenetics and epigenetic drugs |
| Neurodegenerative diseases of motor neurons |
| Amyotrophic lateral sclerosis (ALS) |
| Spinal Muscular Atrophy (SMA) |
| Neurodegenerative Diseases of the Central Nervous System |
| Alzheimer's Disease (AD) |
| Huntington's Disease (HD) |
| Parkinson's Disease (PD) |
| Molecular basis for inheritance |
| DNA and chromosomes |
| Reproduction |
| Recombination and genetic linkage |
| Gene expression |
| Genetic code |
| Gene regulation |
| Genetic change |
| Mutations |
| Natural selection and evolution |
| Medicine |
| Research methods |
| DNA sequencing and genomics |
| Genetic testing: |
| Cell-free fetal DNA |
| Newborn screening |
| Diagnostic testing |
| Carrier testing: |
| Preimplantation genetic diagnosis |
| Prenatal diagnosis |
| Predictive and presymptomatic testing |
| Pharmacogenomics |
| Non-diagnostic testing: |
| Forensic testing |
| Paternity testing |
| Genealogical DNA test |
| Research testing |
| Genome analysis |
| Sequencing |
| Shotgun sequencing |
| High-throughput sequencing |
| Assembly |
| Assembly approaches |
| Finishing |
| Annotation |
| Sequencing pipelines and databases |
| Functional genomics |
| Structural genomics |
| Epigenomics |
| Metagenomics |
| Pharmacogenomics |
| Drug-metabolizing enzymes |
| Predictive prescribing |
| Polypharmacy |
| Drug labeling |
| Mitochondrial genes |
| Replication, repair, transcription and translation |
| Mitochondrial disease |
| Types of genetic disorder: |
| Single-gene |
| Autosomal dominant |
| Autosomal recessive |
| X-linked dominant |
| X-linked recessive |
| Y-linked |
| Mitochondrial |
| Causes of genetic disorder |
| Diagnosis |
| Treatment / gene therapy |
| List of genetic disorder: |
| 1p36 deletion syndrome |
| 18p deletion syndrome |
| 21-hydroxylase deficiency |
| Alpha 1-antitrypsin deficiency |
| AAA syndrome (achalasia-addisonianism-alacrima) |
| Aarskog– Scott syndrome |
| ABCD syndrome |
| Aceruloplasminemia |
| Acheiropodia |
| Achondrogenesis type II |
| Achondroplasia |
| Acute intermittent porphyria |
| Adenylosuccinate lyase deficiency |
| Adrenoleukodystrophy |
| Alagille syndrome |
| Adult syndrome |
| Albinism |
| Alexander disease |
| Alkaptonuria |
| Alport syndrome |
| Alternating hemiplegia of childhood |
| Amyotrophic lateral sclerosis |
| Alström syndrome |
| Alzheimer's disease |
| Amelogenesis imperfecta |
| Aminolevulinic acid dehydratase deficiency porphyria |
| Androgen insensitivity syndrome |
| Angelman syndrome |
| Apert Syndrome |
| Arthrogryposis–renal dysfunction–cholestasis syndrome |
| Ataxia telangiectasia |
| Axenfeld syndrome |
| Beare-Stevenson cutis gyrata syndrome |
| Beckwith–Wiedemann syndrome |
| Benjamin syndrome |
| Biotinidase deficiency |
| Björnstad syndrome |
| Bloom syndrome |
| Birt–Hogg–Dubé syndrome |
| Brody myopathy |
| Cadasil syndrome |
| Carasil syndrome |
| Chronic granulomatous disorder |
| Campomelic dysplasia |
| Canavan disease |
| Carpenter Syndrome |
| Cerebral dysgenesis–neuropathy–ichthyosis–keratoderma syndrome (SEDNIK) |
| Cystic fibrosis |
| Charcot–Marie–Tooth disease |
| CHARGE syndrome |
| Chédiak–Higashi syndrome |
| Cleidocranial dysostosis |
| Cockayne syndrome |
| Coffin–Lowry syndrome |
| Cohen syndrome |
| Collagenopathy, types II and XI |
| Congenital insensitivity to pain with anhidrosis (CIPA) |
| Cowden syndrome |
| CPO deficiency (coproporphyria) |
| Cranio–lenticulo–sutural dysplasia |
| Cri du chat |
| Crohn's disease |
| Crouzon syndrome |
| Crouzonodermoskeletal syndrome (Crouzon syndrome with acanthosis nigricans) |
| Darier's disease |
| Dent's disease (Genetic hypercalciuria) |
| Denys–Drash syndrome |
| De Grouchy syndrome |
| Di George's syndrome |
| Distal hereditary motor neuropathies, multiple types |
| Ehlers–Danlos syndrome |
| Emery–Dreifuss syndrome |
| Erythropoietic protoporphyria |
| Fanconi anemia (FA) |
| Fabry disease |
| Factor V Leiden thrombophilia |
| Familial adenomatous polyposis |
| Familial dysautonomia |
| Feingold syndrome |
| FG syndrome |
| Friedreich's ataxia |
| G6PD deficiency |
| Galactosemia |
| Gaucher disease |
| Gillespie syndrome |
| Griscelli syndrome |
| Hailey-Hailey disease |
| Harlequin type ichthyosis |
| Hemochromatosis, hereditary |
| Hemophilia |
| Hepatoerythropoietic porphyria UROD |
| Hereditary coproporphyria |
| Hereditary hemorrhagic telangiectasia (Osler–Weber–Rendu syndrome) |
| Hereditary Inclusion Body Myopathy |
| Hereditary multiple exostoses |
| Hereditary spastic paraplegia (infantile-onset ascending hereditary spastic paralysis) |
| Hermansky–Pudlak syndrome |
| Hereditary neuropathy with liability to pressure palsies (HNPP) |
| Homocystinuria |
| Huntington's disease |
| Hunter syndrome |
| Hurler syndrome |
| Hutchinson-Gilford progeria syndrome |
| Hyperoxaluria, primary |
| Hyperphenylalaninemia |
| Hypoalphalipoproteinemia (Tangier disease) |
| Hypochondrogenesis |
| Hypochondroplasia |
| Immunodeficiency, centromere instability and facial anomalies syndrome (ICF syndrome) |
| Incontinentia pigmenti |
| Isodicentric 15 |
| Jackson– Weiss syndrome |
| Joubert syndrome |
| Juvenile Primary Lateral Sclerosis (JPLS) |
| Keloid disorder |
| Kniest dysplasia |
| Kosaki overgrowth syndrome |
| Krabbe disease |
| Kufor–Rakeb syndrome |
| LCAT deficiency |
| Lesch-Nyhan syndrome) |
| Li-Fraumeni syndrome |
| Lynch Syndrome |
| Lipoprotein lipase deficiency, familial |
| Marfan syndrome |
| Maroteaux–Lamy syndrome |
| McCune–Albright syndrome |
| McLeod syndrome |
| MEDNIK syndrome |
| Mediterranean fever, familial |
| Menkes disease |
| Methemoglobinemia |
| methylmalonic acidemia |
| Micro syndrome |
| Microcephaly |
| Morquio syndrome |
| Mowat-Wilson syndrome |
| Muenke syndrome |
| Multiple endocrine neoplasia (type 1 and type 2) |
| Muscular dystrophy |
| Muscular dystrophy, Duchenne and Becker type |
| Myostatin-related muscle hypertrophy |
| myotonic dystrophy |
| Natowicz syndrome |
| Neurofibromatosis type I |
| Neurofibromatosis type II |
| Niemann–Pick disease |
| Nonketotic hyperglycinemia |
| nonsyndromic deafness |
| Noonan syndrome |
| Ogden syndrome |
| osteogenesis imperfecta |
| Pantothenate kinase-associated neurodegeneration |
| Patau Syndrome (Trisomy 13) |
| PCC deficiency (propionic acidemia) |
| Porphyria cutanea tarda (PCT) |
| Pendred syndrome |
| Peutz-Jeghers syndrome |
| Pfeiffer syndrome |
| phenylketonuria |
| Pitt–Hopkins syndrome |
| Polycystic kidney disease |
| Polycystic Ovarian Syndrome (PCOS) |
| porphyria |
| Prader-Willi syndrome |
| Primary ciliary dyskinesia (PCD) |
| primary pulmonary hypertension |
| protein C deficiency |
| protein S deficiency |
| Pseudo-Gaucher disease |
| Pseudoxanthoma elasticum |
| Retinitis pigmentosa |
| Rett syndrome |
| Rubinstein-Taybi syndrome (RSTS) |
| Sandhoff disease |
| Sanfilippo syndrome |
| Schwartz–Jampel syndrome |
| spondyloepiphyseal dysplasia congenita (SED) |
| Shprintzen–Goldberg syndrome FBN1 |
| sickle cell anemia |
| Siderius X-linked mental retardation syndrome |
| Sideroblastic anemia |
| Sly syndrome |
| Smith-Lemli-Opitz syndrome |
| Smith Magenis Syndrome |
| Spinal muscular atrophy |
| Spinocerebellar ataxia (types 1-29) |
| SSB syndrome (SADDAN) |
| Stargardt disease (macular degeneration) |
| Stickler syndrome |
| Strudwick syndrome (spondyloepimetaphyseal dysplasia, Strudwick type) |
| Tay-Sachs disease |
| tetrahydrobiopterin deficiency |
| thanatophoric dysplasia |
| Treacher Collins syndrome |
| Tuberous Sclerosis Complex (TSC) |
| Turner syndrome |
| Usher syndrome |
| Variegate porphyria |
| von Hippel-Lindau disease |
| Waardenburg syndrome |
| Weissenbacher-Zweymüller syndrome |
| Williams Syndrome |
| Wilson disease |
| Woodhouse–Sakati syndrome |
| Wolf–Hirschhorn syndrome |
| Xeroderma pigmentosum |
| X-linked mental retardation and macroorchidism (fragile X syndrome) |
| X-linked spinal-bulbar muscle atrophy (spinal and bulbar muscular atrophy) |
| Xp11.22 deletion |
| X-linked severe combined immunodeficiency (X-SCID) |
| X-linked sideroblastic anemia (XLSA) |
| 47,XXX (triple X syndrome) |
| XXXX syndrome (48, XXXX) |
| XXXXX syndrome (49, XXXXX) |
| XYY syndrome (47,XYY) |
| Modern synthesis |
| Four processes |
| Selection |
| Dominance |
| Epistasis |
| Mutation |
| Genetic drift |
| Gene flow |
| Horizontal gene transfer |
| Linkage |
| Applications |
| Explaining levels of genetic variation |
| Detecting selection |
| Demographic inference |
| Evolution of genetic systems |
| Quantitative genetics |
| Genetic epidemiology |
| Statistical genetics |
- Genetic Engineering Topics Topics: 58
- Zoology Topics Topics: 145
- Archaeology Topics Topics: 56
- Charles Darwin Research Topics Topics: 51
- Gene Essay Topics Topics: 77
- Epigenetics Research Topics Topics: 54
- DNA Paper Topics Topics: 113
- Microbiology Paper Topics Topics: 50
- Anatomy Essay Topics Topics: 70
- Biology Topics Topics: 101
- Space Exploration Paper Topics Topics: 76
- Biochemistry Topics Topics: 47
- Atmosphere Paper Topics Topics: 50
- Stem Cell Topics Topics: 100
- Cloning Essay Topics Topics: 74
213 Genetics Research Topics & Essay Questions for College and High School
Genetics studies how genes and traits pass from generation to generation. It has practical applications in many areas, such as genetic engineering, gene therapy, gene editing, and genetic testing. If you’re looking for exciting genetics topics for presentation, you’re at the right place! Here are genetics research paper topics and ideas for different assignments.
🧬 TOP 7 Genetics Topics for Presentation 2024
🏆 best genetics essay topics, ❓ genetics research questions, 👍 good genetics research topics & essay examples, 🌟 cool genetics topics for presentation, 🌶️ hot genetics topics to write about, 🔎 current genetic research topics, 🎓 most interesting genetics topics.
- Advantages and Disadvantages of Genetic Testing
- Should Parents Have the Right to Choose Their Children Based on Genetics?
- Genetic and Social Behavioral Learning Theories
- Genetically Modified Pineapples and Their Benefits
- Genomics, Genetics, and Nursing Involvement
- Link Between Obesity and Genetics
- The Importance of Heredity and Genetics
- Mendelian Genetics and Chlorophyll in Plants This paper investigates Mendelian genetics. This lab report will examine the importance of chlorophyll in plants using fast plants’ leaves and stems.
- Genetic and Environmental Impacts on Teaching Work If students do not adopt learning materials and the fundamentals of the curriculum well, this is a reason for reviewing the current educational regimen.
- Isolated by Genetics but Longing to Belong The objective of this paper is to argue for people with genetic illnesses to be recognized and appreciated as personages in all institutions.
- The Concept of Epigenetics Epigenetics is a study of heritable phenotypic changes or gene expression in cells that are caused by mechanisms other than DNA sequence.
- Op-ED Genetic Engineering: The Viewpoint The debate about genetic engineering was started more than twenty years ago and since that time it has not been resolved
- The Potential Benefits of Genetic Engineering Genetic engineering is a new step in the development of the humans’ knowledge about the nature that has a lot of advantages for people in spite of its controversial character.
- Aspects of the Genetic Diseases Genetic diseases are disorders that happen through mutations that occur in the human body. They can be monogenic, multifactorial, and chromosomal.
- Genetic Diseases: Hemophilia This article focuses on a genetic disorder such as hemophilia: causes, symptoms, history, diagnosis, and treatment.
- Genetics: Gaucher Disease Type 1 The Gaucher disease type 1 category is a genetically related complication in which there is an automatic recession in the way lysosomes store some important gene enzymes.
- Genetic Disorders: Diagnosis, Screening, and Treatment Chorionic villus is a test of sampling done especially at the early stages of pregnancy and is used to identify some problems which might occur to the fetus.
- Research of Genetic Disorders Types This essay describes different genetic disorders such as hemophilia, turner syndrome and sickle cell disease (SCD).
- Relation Between Genetics and Intelligence Intelligence is a mental ability to learn from experience, tackle issues and use knowledge to adapt to new situations and the factor g may access intelligence of a person.
- Type 1 Diabetes in Children: Genetic and Environmental Factors The prevalence rate of type 1 diabetes in children raises the question of the role of genetic and environmental factors in the increasing cases of this illness.
- Human Genetics: Multifactorial Traits This essay states that multifactorial traits in human beings are essential for distinguishing individual characteristics in a population.
- What Is Silencer Rna in Genetics RNA silencing is an evolutionary conserved intracellular surveillance system based on recognition. RNA silencing is induced by double-stranded RNA sensed by the enzyme Dicer.
- GMO Use in Brazil and Other Countries The introduction of biotechnology into food production was a milestone. Brazil is one of the countries that are increasingly using GMOs for food production.
- Genetically Modified Food Safety and Benefits Today’s world faces a problem of the shortage of food supplies to feed its growing population. The adoption of GM foods can solve the problem of food shortage in several ways.
- Genetically Modified Organisms: Pros and Cons Genetically modified organisms are organisms that are created after combining DNA from a different species into an organism to come up with a transgenic organism.
- Genetics and Evolution: Mutation, Selection, Gene Flow and Drift Evolutionary genetics deals with mechanisms that explain the presence and maintenance of traits responsible for genetic variations.
- Exploring ADHD: Genetics, Environment, and Brain Changes Attention deficit hyperactivity disorder is the most prevalent child behavioral disorder characterized by inattention, hyperactivity, and impulsivity.
- Down’s Syndrome as a Genetic Disorder Many people are born with genetic diseases that manifest themselves in one way or another throughout their lives. One of these abnormalities is Down’s syndrome.
- Addiction: Genetic, Environmental, and Psychological Factors Addiction: the role of dopamine and its impact on the brain’s reward system exacerbates addiction and highlights the need for a comprehensive approach.
- Procreative Beneficence: Technological Developments in Genetics Technological developments in genetics have revolutionized procreation by allowing parents to choose the most intelligent genes for their offspring.
- Genetic Technologies for Pathogen Identification The paper states that a genotype represents a set of genes and determines the organism’s phenotype by promoting the development of certain traits.
- Epigenetics as the Phenomenon and Its Examples Epigenetics, or epigenomics, is the study of how the expression of genes that do not presuppose irreversible alterations in the underlying DNA sequence changes.
- Genetics: When Nurture Becomes Nature The paper aims to review the environmental and dietary aspects of epigenetics and show how the research can be useful in understanding genetics.
- Is ADHD Genetically Passed Down to Family Members? Genetic correlations between such qualities as hyperactivity and inattention allowed us to define ADHD as a spectrum disorder rather than a unitary one.
- Alzheimer’s Disease: Genetic Risk and Ethical Considerations Alzheimer’s disease is a neurodegenerative disease that causes brain shrinkage and the death of brain cells. It is the most prevalent form of dementia.
- Behavioral Genetics in “Harry Potter” Books The reverberations of the Theory of Behavioral Genetics permeate the Harry Potter book series, enabling to achieve the comprehension of characters and their behaviors.
- Environmental Impact of Genetically Modified Crop In 1996, the commercial use of genetically modified (GM) crop production techniques had increasingly been accepted by many farmers.
- Gene Transfer and Genetic Engineering Mechanisms This paper discusses gene transfer mechanisms and the different genetic engineering mechanisms. Gene transfer, a natural process, can cause variation in biological features.
- Nutrition: Obesity Pandemic and Genetic Code The environment in which we access the food we consume has changed. Unhealthy foods are cheaper, and there is no motivation to eat healthily.
- Genetics in Diagnosis of Diseases Medical genetics aims to study the role of genetic factors in the etiology and pathogenesis of various human diseases.
- How Much Do Genetics Affect Us?
- What Can Livestock Breeders Learn From Conservation Genetics and Vice Versa?
- How Do Genetics Affect Caffeine Tolerance?
- How Dolly Sheep Changed Genetics Forever?
- What Is the Nature and Function of Genetics?
- What Are the Five Branches of Genetics?
- How Does Genetics Affect the Achievement of Food Security?
- Are Owls and Larks Different in Genetics When It Comes to Aggression?
- How Do Neuroscience and Behavioral Genetics Improve Psychiatric Assessment?
- How Does Genetics Influence Human Behavior?
- What Are Three Common Genetics Disorders?
- Can Genetics Cause Crime or Are We Presupposed?
- What Are Examples of Genetics Influences?
- How Do Genetics Influence Psychology?
- What Traits Are Influenced by Genetics?
- Why Tampering With Our Genetics Will Be Beneficial?
- How Genetics and Environment Affect a Child’s Behaviors?
- Which Country Is Best for Genetics Studies?
- How Does the Environment Change Genetics?
- Can Crop Models Identify Critical Gaps in Genetics, Environment, and Management Interactions?
- How Can Drug Metabolism and Transporter Genetics Inform Psychotropic Prescribing?
- Can You Change Your Genetics?
- How Old Are European Genetics?
- Will Benchtop Sequencers Resolve the Sequencing Trade-off in Plant Genetics?
- What Can You Study in Genetics?
- What Are Some Genetic Issues?
- Does Genetics Matter for Disease-Related Stigma?
- How Did the Drosophila Melanogaster Impact Genetics?
- What Is a Genetics Specialist?
- Will Genetics Destroy Sports?
- The Morality of Selective Abortion and Genetic Screening The paper states that the morality of selective abortion and genetic screening is relative. This technology should be made available and legal.
- Environmental Ethics in Genetically Modified Organisms The paper discusses genetically modified organisms. Environmental ethics is centered on the ethical dilemmas arising from human interaction with the nonhuman domain.
- Does Genetic Predisposition Affect Learning in Other Disciplines? This paper aims to examine each person’s ability to study a discipline for which there is no genetic ability and to understand how effective it is.
- Cause and Effect of Genetically Modified Food The paper states that better testing should be done on GMOs. It would lead to avoiding catastrophic health issues caused by these foods.
- Detection of Genetically Modified Products Today, people are becoming more concerned about the need to protect themselves from the effects of harmful factors and to buy quality food.
- Genetically Modified Organisms Solution to Global Hunger It is time for the nations to work together and solve the great challenge of feeding the population by producing sufficient food and using fewer inputs.
- Genetic Engineering: Cloning With Pet-28A Embedding genes into plasmid vectors is an integral part of molecular cloning as part of genetic engineering. An example is the cloning of the pectate lyase gene.
- Restricting the Volume of Sale of Fast Foods and Genetically Modified Foods The effects of fast foods and genetically modified foods on the health of Arizona citizens are catastrophic. The control of such outlets and businesses is crucial.
- Researching of Genetic Engineering DNA technology entails the sequencing, evaluation and cut-and-paste of DNA. The following paper analyzes the historical developments, techniques, applications, and controversies.
- Genetically Modified Crops: Impact on Human Health The aim of this paper is to provide some information about genetically modified crops as well as highlight the negative impacts of genetically modified soybeans on human health.
- Genetic Engineering Biomedical Ethics Perspectives Diverse perspectives ensure vivisection, bio, and genetic engineering activities, trying to deduce their significance in evolution, medicine, and society.
- Down Syndrome: The Genetic Disorder Down syndrome is the result of a glandular or chemical disbalance in the mother at the time of gestation and of nothing else whatsoever.
- Genetic Modifications: Advantages and Disadvantages Genetic modifications of fruits and vegetables played an important role in the improvement process of crops and their disease resistance, yields, eating quality and shelf life.
- Genetics of Personality Disorders The genetics of different psychological disorders can vary immensely; for example, the genetic architecture of schizophrenia is quite perplexing and complex.
- Labeling of Genetically Modified Products Regardless of the reasoning behind the labeling issue, it is ethical and good to label the food as obtained from genetically modified ingredients for the sake of the consumers.
- Convergent Evolution, Genetics and Related Structures This paper discusses the concept of convergent evolution and related structures. Convergent evolution describes the emergence of analogous or similar traits in different species.
- Genetic Technologies in the Healthcare One area where genetic technology using DNA works for the benefit of society is medicine, as it will improve the treatment and management of genetic diseases.
- Are Genetically Modified Organisms Really That Bad? Almost any food can be genetically modified: meat, fruits, vegetables, etc. Many people argue that consuming products, which have GMOs may cause severe health issues.
- Discussion of Genetic Testing Aspects The primary aim of the adoption process is to ensure that the children move into a safe and loving environment.
- Ethical Concerns on Genetic Engineering The paper discusses Clustered Regularly Interspaced Short Palindromic Repeats technology. It is a biological system for modifying DNA.
- The Normal Aging Process and Its Genetic Basis Various factors can cause some genetic disorders linked to premature aging. The purpose of this paper is to talk about the genetic basis of the normal aging process.
- Medicine Is Not a Genetic Supermarket Together with the development of society, medicine also develops, but some people are not ready to accept everything that science creates.
- Epigenetics: Definition and Family History Epigenetics refers to the learning of fluctuations in creatures induced by gene expression alteration instead of modification of the ‘genetic code itself.
- Genetically Modified Organisms in Aquaculture Genetically Modified Organisms are increasingly being used in aquaculture. They possess a unique genetic combination that makes them uniquely suited to their environment.
- Genetic Modification of Organisms to Meet Human Needs Genetic modification of plants and animals for food has increased crop yields as the modified plants and animals have more desirable features such as better production.
- Discussion of Epigenetics Meanings and Aspects The paper discusses epigenetics – the study of how gene expression takes place without changing the sequence of DNA.
- Genetically Modified Products: Positive and Negative Sides This paper considers GMOs a positive trend in human development due to their innovativeness and helpfulness in many areas of life, even though GMOs are fatal for many insects.
- Overview of African Americans’ Genetic Diseases African Americans are more likely to suffer from certain diseases than white Americans, according to numerous studies.
- Plant Genetic Engineering: Genetic Modification Genetic engineering is the manipulation of the genes of an organism by completely altering the structure of the organism.
- Genetically Modified Fish: The Threats and Benefits This article’s purpose is to evaluate possible harm and advantages of genetically modified fish. For example, the GM fish can increase farms’ yield.
- DNA and the Birth of Molecular Genetics Molecular genetics is critical in studying traits that are passed through generations. The paper analyzes the role of DNA to provide an ample understanding of molecular genetics.
- Natural Selection and Genetic Variation The difference in the genetic content of organisms is indicative that certain group of organisms will stay alive, and effectively reproduce than other organisms residing in the same environment.
- Genetically Modified Foods: How Safe are they? This paper seeks to address the question of whether genetically modified plants meant for food production confer a threat to human health and the environment.
- The Genetic Material Sequencing This experiment is aimed at understanding the real mechanism involved in genetic material sequencing through nucleic acid hybridization.
- Genetically Modified Organisms in Human Food This article focuses on Genetically Modified Organisms as they are used to produce human food in the contemporary world.
- Genetic Disorder Cystic Fibrosis Cystic fibrosis is a genetic disorder. The clinical presentation of the disease is evident in various organs of the body as discussed in this paper.
- Human Genome and Application of Genetic Variations Human genome refers to the information contained in human genes. The Human Genome Project (HGP) focused on understanding genomic information stored in the human DNA.
- Saudi Classic Aniridia Genetic and Genomic Analysis This research was conducted in Saudi Arabia to determine the genetic and genomic alterations that underlie classic anirida.
- What Makes Humans Mortal Genetically? The causes of aging have been studied and debated about by various experts for centuries, there multiple views and ideas about the reasons of aging and.
- Decision Tree Analysis and Genetic Algorithm Methods Application in Healthcare The paper investigates the application of such methods of data mining as decision tree analysis and genetic algorithm in the healthcare setting.
- The role of genes in our food preferences.
- The molecular mechanisms of aging and longevity.
- Genomic privacy: ways to protect genetic information.
- The effects of genes on athletic performance.
- CRISPR-Cas9 gene editing: current applications and future perspectives.
- Genetic underpinnings of human intelligence.
- The genetic foundations of human behavior.
- The role of DNA analysis in criminal justice.
- The influence of genetic diversity on a species’ fate.
- Genetic ancestry testing: the process and importance.
- Ban on Genetically Modified Foods Genetically modified (GM) foods are those that are produced with the help of genetic engineering. Such foods are created from organisms with changed DNA.
- Genetic Screening and Testing The provided descriptive report explains how genetic screening and testing assists clinicians in determining cognitive disabilities in babies.
- Neurobiology: Epigenetics in Cocaine Addiction Studies have shown that the addiction process is the interplay of many factors that result in structural modifications of neuronal pathways.
- Family Pedigree, Human Traits, and Genetic Testing Genetic testing allows couples to define any severe genes in eight-cell embryos and might avoid implanting the highest risk-rated ones.
- Darwin’s Theory of Evolution: Impact of Genetics New research proved that genetics are the driving force of evolution which causes the revision of some of Darwin’s discoveries.
- Genetic Tests: Pros and Cons Genetic testing is still undergoing transformations and further improvements, so it may be safer to avoid such procedures under certain circumstances.
- Case on Preserving Genetic Mutations in IVF In the case, a couple of a man and women want to be referred to an infertility specialist to have a procedure of in vitro fertilization (IVF).
- Race: Genetic or Social Construction One of the most challenging questions the community faces today is the following: whether races were created by nature or society or not.
- Huntington’s Chorea Disease: Genetics, Symptoms, and Treatment Huntington’s chorea disease is a neurodegenerative heritable disease of the central nervous system that is eventually leading to uncontrollable body movements and dementia.
- Genetics: A Frameshift Mutation in Human MC4R This article reviews the article “A Frameshift Mutation in Human mc4r Is Associated With Dominant Form of Obesity” published by C. Vaisse, K. Clement, B. Guy-Grand & P. Froguel.
- DNA Profiling: Genetic Variation in DNA Sequences The paper aims to determine the importance of genetic variation in sequences in DNA profiling using specific techniques.
- Benefits of Genetic Engineering The potential increase of people’s physical characteristics and lifespan may be regarded as another advantage of genetic engineering.
- Simulating the Natural Selection and Genetic Drift This lab was aimed at simulating the natural selection and genetic drift as well as predicting their frequency of evolution change.
- Cystic Fibrosis: Genetic Disorder Cystic fibrosis, also referred to as CF, is a genetic disorder that can affect the respiratory and digestive systems.
- Genetic Testing and Privacy & Discrimination Issues Genetic testing is fraught with the violation of privacy and may result in discrimination in employment, poor access to healthcare services, and social censure.
- Genetics or New Pharmaceutical Article Within the Last Year Copy number variations (CNVs) have more impacts on DNA sequence within the human genome than single nucleotide polymorphisms (SNPs).
- Genetic Mechanism of Colorectal Cancer Colorectal Cancer (CRC) occurrence is connected to environmental factors, hereditary factors, and individual ones.
- Genetic Association and the Prognosis of Phenotypic Characters The article understudy is devoted to the topic of genetic association and the prognosis of phenotypic characters. The study focuses on such a topic as human iris pigmentation.
- PiggyBac Transposon System in Genetics Ideal delivery systems for gene therapy should be safe and efficient. PB has a high transposition efficiency, stability, and mutagenic potential in most mammalian cell lines.
- Technology of Synthesis of Genetically Modified Insulin The work summarizes the technology for obtaining genetically modified insulin by manipulating the E. coli genome.
- A Career in Genetics: Required Skills and Knowledge A few decades ago, genetics was mostly a science-related sphere of employment. People with a degree in genetics can have solid career prospects in medicine and even agriculture.
- Advantages of Using Genetically Modified Foods Genetic modifications of traditional crops have allowed the expansion of agricultural land in areas with adverse conditions.
- Genetic Factors as the Cause of Anorexia Nervosa Genetic predisposition currently seems the most plausible explanation among all the proposed etiologies of anorexia.
- Personality Is Inherited Principles of Genetics The present articles discusses the principles of genetics, and how is human temperament and personality formed.
- Literature Review: Acceptability of Genetic Engineering The risks and benefits of genetic engineering must be objectively evaluated so that modern community could have a better understanding of this problem
- Impacts of Genetic Engineering of Agricultural Crops In present days the importance of genetic engineering grew due to the innovations in biotechnologies and Sciences.
- The Effects of Genetic Modification of Agricultural Products Discussion of the threat to the health of the global population of genetically modified food in the works of Such authors as Jane Brody and David Ehrenfeld.
- Genetic Engineering in Food and Freshwater Issues The technology of bioengineered foods, genetically modified, genetically engineered, or transgenic crops, will be an essential element in meeting the challenging population needs.
- Genetic Engineering and Religion: Designer Babies The current Pope has opposed any scientific procedure, including genetic engineering, in vitro fertilization, and diagnostic tests to see if babies have disabilities.
- Genetically Modified Food as a Current Issue GM foods are those kinds of food items that have had their DNA changed by usual breeding; this process is also referred to as Genetic Engineering.
- All About the Role of Genetic Engineering and Biopiracy The argument whether genetically engineered seeds have monopolized the market in place of the contemporary seeds has been going on for some time now.
- Genetic Engineering and Cloning Controversy Genetic engineering and cloning are the most controversial issues in modern science. The benefits of cloning are the possibility to treat incurable diseases and increase longevity.
- Biotechnology: Methodology in Basic Genetics The material illustrates the possibilities of ecological genetics, the development of eco-genetical models, based on the usage of species linked by food chain as consumers and producers.
- Genetics Impact on Health Care in the Aging Population This paper briefly assesses the impact that genetics and genomics can have on health care costs and services for geriatric patients.
- Concerns Regarding Genetically Modified Food It is evident that genetically modified food and crops are potentially harmful. Both humans and the environment are affected by consequences as a result of their introduction.
- Family Genetic History and Planning for Future Wellness The patient has a family genetic history of cardiac arrhythmia, allergy, and obesity. These diseases might lead to heart attacks, destroy the cartilage and tissue around the joint.
- Genetic Predisposition to Alcohol Dependence and Alcohol-Related Diseases The subject of genetics in alcohol dependence deserves additional research in order to provide accurate results.
- Genetically-Modified Fruits, Pesticides, or Biocontrol? The main criticism of GMO foods is the lack of complete control and understanding behind GMO processes in relation to human consumption and long-term effects on human DNA.
- Genetic Variants Influencing Effectiveness of Exercise Training Programmes “Genetic Variants Influencing Effectiveness of Exercise Training Programmes” studies the influence of most common genetic markers that indicate a predisposition towards obesity.
- Eugenics, Human Genetics and Their Societal Impact Ever since the discovery of DNA and the ability to manipulate it, genetics research has remained one of the most controversial scientific topics of the 21st century.
- Genetic foundations of rare diseases.
- Genetic risk factors for neurodegenerative disorders.
- Inherited cancer genes and their impact on tumor development.
- Genetic variability in drug metabolism and its consequences.
- The role of genetic and environmental factors in disease development.
- Genomic cancer medicine: therapies based on tumor DNA sequencing.
- Non-invasive prenatal testing: benefits and challenges.
- Genetic basis of addiction.
- The origins of domestication genes in animals.
- How can genetics affect a person’s injury susceptibility?
- Genetic Interference in Caenorhabditis Elegans The researchers found out that the double-stranded RNA’s impact was not only the cells, it was also on the offspring of the infected animals.
- Genetics and Autism Development Autism is associated with a person’s genetic makeup. This paper gives a detailed analysis of this condition and the role of genetics in its development.
- Start Up Company: Genetically Modified Foods in China The aim of establishing the start up company is to develop the scientific idea of increasing food production using scientific methods.
- Community Health Status: Development, Gender, Genetics Stage of development, gender and genetics appear to be the chief factors that influence the health status of the community.
- Genetics of Developmental Disabilities The aim of the essay is to explore the genetic causes of DDs, especially dyslexia, and the effectiveness of DNA modification in the treatment of these disorders.
- Homosexuality as a Genetic Characteristic The debate about whether homosexuality is an inherent or social parameter can be deemed as one of the most thoroughly discussed issues in the contemporary society.
- Autism Spectrum Disorder in Twins: Genetics Study Autism spectrum disorder is a behavioral condition caused by genetic and environmental factors. Twin studies have been used to explain the hereditary nature of this condition.
- Why Is the Concept of Epigenetics so Fascinating? Epigenetics has come forward to play a significant role in the modern vision of the origin of illnesses and methods of their treatment, which results in proving to be fascinating.
- Epigenetics and Its Effect on Physical and Mental Health This paper reviews a research article and two videos on epigenetics to developing an understanding of the phenomenon and how it affects individuals’ physical and mental health.
- Genetic Counseling for Cystic Fibrosis Some of the inherited genes may predispose individuals to specific health conditions like cystic fibrosis, among other inheritable diseases.
- Genetic and Genomic Healthcare: Nurses Ethical Issues Genomic medicine is one of the most significant ways of tailoring healthcare at a personal level. This paper will explore nursing ethics concerning genetic information.
- Medical and Psychological Genetic Counseling Genetic counseling is defined as the process of helping people understand and adapt to the medical, psychological, and familial implications of genetic contributions to disease.
- Patent on Genetic Discoveries and Supreme Court Decision Supreme Court did not recognize the eligibility of patenting Myriad Genetics discoveries due to the natural existence of the phenomenon.
- Genetic Testing, Its Background and Policy Issues This paper will explore the societal impacts of genetic research and its perceptions in mass media, providing argumentation for support and opposition to the topic.
- Genetically Modified Organisms and Future Farming There are many debates about benefits and limitations of GMOs, but so far, scientists fail to prove that the advantages of these organisms are more numerous than the disadvantages.
- GMO: Some Peculiarities and Associated Concerns Genetically modified organisms are created through the insertion of genes of other species into their genetic codes.
- Mitosis, Meiosis, and Genetic Variation According to Mendel’s law of independent assortment, alleles for different characteristics are passed independently from each other.
- Genetic Counseling and Hypertension Risks This paper dwells upon the peculiarities of genetic counseling provided to people who are at risk of developing hypertension.
- The Perspectives of Genetic Engineering in Various Fields Genetic engineering can be discussed as having such potential benefits for the mankind as improvement of agricultural processes, environmental protection, resolution of the food problem.
- Labeling Food With Genetically Modified Organisms The wide public has been concerned about the issue of whether food products with genetically modified organisms should be labeled since the beginning of arguments on implications.
- Diabetes Genetic Risks in Diagnostics The introduction of the generic risks score in the diagnosis of diabetes has a high potential for use in the correct classification based on a particular type of diabetes.
- Residence and Genetic Predisposition to Diseases The study on the genetic predisposition of people to certain diseases based on their residence places emphasizes the influence of heredity.
- Controversies in Genetics: Eugenics, Ethics, and Human Health Ethical issues associated with human genetics and eugenics have been recently brought to public attention, resulting in the creation of peculiar public policy.
- Value of the Epigenetics Epigenetics is a quickly developing field of science that has proven to be practical in medicine. It focuses on changes in gene activity that are not a result of DNA sequence mutations.
- Genetics Seminar: The Importance of Dna Roles DNA has to be stable. In general, its stability becomes possible due to a large number of hydrogen bonds which make DNA strands more stable.
- Genetically Modified Organisms: Position Against Genetically modified organisms are organisms that are created after combining DNAs of different species to come up with a transgenic organism.
- GMOs: Enhancing Food Security and Nutrition Scientists believe GMOs can feed everyone in the world. This can be achieved if governments embrace the use of this new technology to create genetically modified foods.
- WHO’s Stance on Genetically Modified Foods and Global Reactions Genetically modified foods have elicited different reactions all over the world with some countries banning its use while others like the United States allowing its consumption.
- The Role of Genetics in Health and Personality Traits The branch of biology that deals with variation, heredity, and their transmission in both animals and the plant is called genetics.
- Genetic Engineering: Gene Therapy The purpose of the present study is to discover just what benefits gene therapy might have to offer present and future generations.
- Genetically Modified Foods and Their Impact on Human Health Genetically modified food has become the subject of discussion. There are numerous benefits and risks tied to consumption of genetically modified foods.
- Genetic Engineering: Dangers and Opportunities Genetic engineering can be defined as: “An artificial modification of the genetic code of an organism. It changes radically the physical nature of the being in question.
Cite this post
- Chicago (N-B)
- Chicago (A-D)
StudyCorgi. (2022, January 16). 213 Genetics Research Topics & Essay Questions for College and High School. https://studycorgi.com/ideas/genetics-essay-topics/
"213 Genetics Research Topics & Essay Questions for College and High School." StudyCorgi , 16 Jan. 2022, studycorgi.com/ideas/genetics-essay-topics/.
StudyCorgi . (2022) '213 Genetics Research Topics & Essay Questions for College and High School'. 16 January.
1. StudyCorgi . "213 Genetics Research Topics & Essay Questions for College and High School." January 16, 2022. https://studycorgi.com/ideas/genetics-essay-topics/.
Bibliography
StudyCorgi . "213 Genetics Research Topics & Essay Questions for College and High School." January 16, 2022. https://studycorgi.com/ideas/genetics-essay-topics/.
StudyCorgi . 2022. "213 Genetics Research Topics & Essay Questions for College and High School." January 16, 2022. https://studycorgi.com/ideas/genetics-essay-topics/.
These essay examples and topics on Genetics were carefully selected by the StudyCorgi editorial team. They meet our highest standards in terms of grammar, punctuation, style, and fact accuracy. Please ensure you properly reference the materials if you’re using them to write your assignment.
This essay topic collection was updated on June 22, 2024 .
- Open access
- Published: 23 November 2021
Genetic/genomic testing: defining the parameters for ethical, legal and social implications (ELSI)
- Tania Ascencio-Carbajal 1 ,
- Garbiñe Saruwatari-Zavala 3 ,
- Fernando Navarro-Garcia 1 , 2 &
- Eugenio Frixione 1 , 2
BMC Medical Ethics volume 22 , Article number: 156 ( 2021 ) Cite this article
8875 Accesses
7 Citations
8 Altmetric
Metrics details
Genetic/genomic testing (GGT) are useful tools for improving health and preventing diseases. Still, since GGT deals with sensitive personal information that could significantly impact a patient’s life or that of their family, it becomes imperative to consider Ethical, Legal and Social Implications (ELSI). Thus, ELSI studies aim to identify and address concerns raised by genomic research that could affect individuals, their family, and society. However, there are quantitative and qualitative discrepancies in the literature to describe the elements that provide content to the ELSI studies and such problems may result in patient misinformation and harmful choices.
We analyzed the major international documents published by international organizations to specify the parameters that define ELSI and the recognized criteria for GGT, which may prove useful for researchers, health professionals and policymakers. First, we defined the parameters of the ethical, legal and social fields in GGT to avoid ambiguities when using the acronym ELSI. Then, we selected nine documents from 44 relevant publications by international organizations related to genomic medicine.
We identified 29 ELSI sub-criteria concerning to GGT, which were organized and grouped within 10 minimum criteria: two from the ethical field, four from the legal field and four from the social field. An additional analysis of the number of appearances of these 29 sub-criteria in the analyzed documents allowed us to order them and to determine 7 priority criteria for starting to evaluate and propose national regulations for GGT.
Conclusions
We propose that the ELSI criteria identified herein could serve as a starting point to formulate national regulation on personalized genomic medicine, ensuring consistency with international bioethical requirements.
Peer Review reports
Genetic/genomic testing (GGT) refers to complementary tools used for improving health and preventing diseases. Genetic testing detects specific mutations in the genome of a patient for identifying monogenic diseases. In contrast, genomic testing detects risk factors and predisposition to diseases involving more than one gene [ 1 ]. Thus, GGT can identify features in the DNA of a patient that may affect her/his health, helping physicians to (a) prevent or at least delay the appearance of resulting illnesses, (b) estimate disease risk for family members, and (c) avoid the risk of transmitting those risks to descendants [ 2 , 3 ]. Since GGT deals with such sensitive personal information, which could significantly impact the life of a patient or their families, it becomes imperative taking into account ethical, legal, and social considerations when practicing it [ 4 ]. Key considerations are still not entirely well defined.
In order to address these issues, Ethical, Legal, and Social Implications (ELSI) studies were formally started in 1990 as part of the Human Genome Project, with an aim at identifying and confronting the troubles posed by genomic research that could affect individuals, their family members and eventually society at large. ELSI studies are today an interdisciplinary research area in constant evolution and expansion, currently embracing much more than intended at its beginnings 30 years ago. The complex connections among ethical, legal and social studies have resulted in the term ELSI being commonly understood as an integral set instead of an aggregate of independent elements, thus turning it into a somewhat fuzzy entity [ 5 ]. Additionally, there are quantitative and qualitative discrepancies in the literature when describing the elements that provide content to each field of ELSI studies regarding to genomic and genetic testing as well as public health on genomics and genetics [ 4 , 6 , 7 ]. These discrepancies are also notice in international documents published by international organizations. There is no clear distinction as to whether the elements of study addressed belong to the ethical, legal or social field; sometimes they are only cited as ELSI [ 8 , 9 , 10 ], or they are referred to only as ethical principles although they include the legal and social field [ 11 , 12 ]. In addition, there are differences in how many and which ELSI criteria they belong [ 8 , 13 ]. This lack of agreement in the elements that define ELSI, as well as in the criteria linked to the information that arises from the practice of GGT, can generate confusion in policy and decision makers, who may lose sight of the relevance or even the urgency of addressing certain issues, leading to difficulties in developing regulations with international equivalences regarding the use of genomic technology and hindering international scientific cooperation. These inaccurate policies and decisions may end up affecting the rights of patients (i.e. government decisions for implementing and expanding newborn screening programs that impact on children rights to health [ 14 ]). In some cases, autonomy may be affected, such as the case of the Havasupai Indian Tribe where the right to informed consent and to know or not the results of the tests was violated [ 15 ]. As well as lead patients to make harmful health choices (i.e. patients who do not seek prompt treatment due to a false negative result on direct-to-consumer testing (DTC) for the detection of the BRCA gene [ 16 ]. In addition, the lack of agreement on privacy issues may open the field to (a) leave personal genetic data of users unprotected, and therefore exposed to violation of their privacy and that of their families; and (b) misuse of the genomic information of a person by third parties with economic, health or discrimination consequences, among other hazards [ 6 , 17 , 18 ].
There have been significant efforts to address these issues [ 10 , 19 , 20 , 21 , 22 ]. However, some of them have been biased towards the clinical research part, setting aside other essential ELSI criteria for GGT such as those of commercialization and health regulation criteria, making it difficult to define parameters to include particular aspects for GGT within the concepts of ELSI. Consequently, quantitative and qualitative inconsistencies remain in the official and research literatures, which hamper a uniform description of the elements that provide content to each ELSI study field and ELSI criteria. In this work, we focus on defining the ethical, legal and social implications (ELSI) for genetic/genomic testing (GGT), but to achieve this goal, we first specified the parameters that define the ELSI fields. This main goal was achieved through an analysis of the major international documents on genomic medicine published by international organizations. Our analysis and the data generated may be useful for researchers, healthcare professionals and policymakers as an unbiased, synthesized, comprehensive view of relevant ELSI topics.
In general, this study is limited to the ELSI aspects linked to GGT tests being carried out for disease detection or estimating the risk of developing one in adults with full consent capacity. In order to find out what exactly are the parameters that define ELSI criteria associated with GGT, we analyzed the documents published by international organizations related to ELSI subjects by following an adapted version of the steps recommended by Strech and Sofaer regarding a systematic review of reason [ 23 ], see also Fig. 1 . This is because according to Boyle 1999, international documents are guides that help countries to regulate their own practices within, in this way soft law allows to create non-binding guidelines that becomes national binding regulations [ 24 ]. Additionally, in this work, we use the word "documents" in a sense that encompasses both instruments (binding for the signatory countries and non-binding) as well as recommendations made by panels of experts coordinated by international organizations. Thus, the international documents related to ELSI were subjected to the Strech and Sofaer systematic review based on four steps: (1) formulation of the review question and eligibility criteria, (2) selection of all the documents that applies to the criteria, (3) extraction and synthesis of information, (4) presenting the results with an answer to the review question.
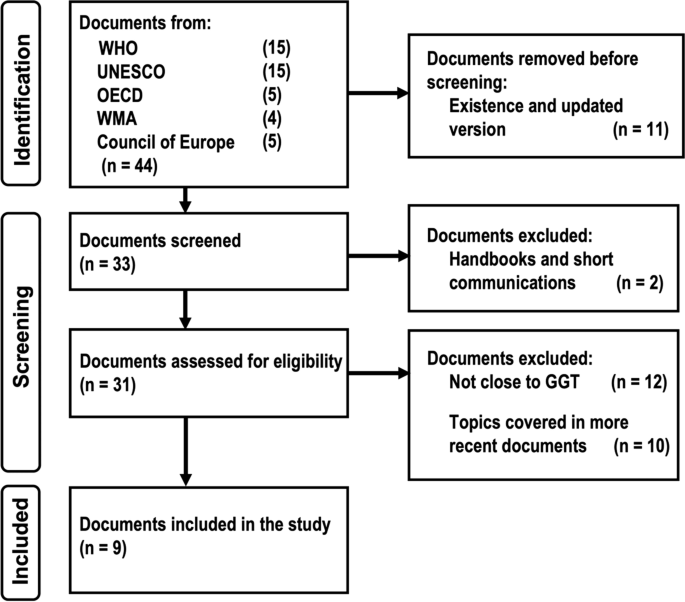
Flow diagram for selecting GGT-related content in databases of major international ELSI documents
The study question was defined as: What are the main ELSI criteria which international organizations have demanded for GGT over the last thirty years? Eligibility criteria were set as: documents published by international organization in the last 30 years, related to GGT tests for detection or estimating risk of diseases in adults with full consent capacity. To address the second step of the systematic review, we carried out a systematic search in databases of the major international organizations related to genomic medicine, mainly focusing on guidelines that could work for different countries rather than on particularities, according to Boyle (1999) as mentioned above. To identify the international documents concerning GGT, we carried out a systematic search of the terms “genomic medicine," "genetic testing" and "human rights and health" in databases of the major international organizations related to genomic medicine: World Health Organization (WHO), World Medical Association (WMA), Organization for Economic Co-operation and Development (OECD), United Nations Educational, Scientific and Cultural Organization (UNESCO), and Council of Europe. After several testing, these general key words were selected in order to minimize the exclusion of relevant documents, and for covering as much as possible by minimizing the bias that would leave some documents out. Next, the records of 30 years to date related to genomic medicine for health purposes in adults with full consent capacity were selected. Figure 1 shows the flow diagram for the identification of literature about ELSI of GGT and the result of it according to page et al. [ 25 ].
For the third step on extraction and synthesis of information, we carried out a systematic and detailed scrutiny of the identified documents. Each entire document was read to identify core concepts that met eligibility criteria in each article or statement. Final core concepts identified were reviewed, agreed and approved by all authors (differences were settled by majority): informed consent, non-discrimination, counseling, privacy and confidentiality issues, regulation, equity and accessibility, quality, trained medical personnel. A subsequent screening was performed to identify ideas associated with core concepts by either textual content or semantic ideas, and wrote it down as a "criterion" in a list. Finally, for the fourth step, these criteria were grouped by thematic affinity and thus defined as criteria and sub-criteria. Then these criteria and sub-criteria were assigned, by common agreement between authors, to the corresponding ETHICAL, LEGAL and SOCIAL field according to the definitions for fields indicated below.
Primary criteria in terms of coverage priority were then selected to be considered in regulations oriented to protecting all personal genetic information and rights. Finally, all data are discussed from present and future perspectives.
Defining ELSI parameters
Defining the parameters for the ethical, legal and social fields is essential to avoid ambiguities when using the acronym ELSI. In order to address GGT criteria, first the ELSI fields were clarified. Although exist different definitions to differentiate ELSI fields [ 20 , 26 ], we build along “ethical, legal and social issues in science and technology” guidelines proposed by Chameau et al. [ 27 ], because they clearly identify and separate ethical, legal and social fields. In addition, human rights themselves set guidelines on ethical principles for treating persons and therefore a minimum of universal standards for rights of patients [ 28 ]. Among universal ethical principles, those established by Beauchamp and Childress [ 29 ] (beneficence, non-maleficence, autonomy and justice) have resulted in a common framework for medical practice [ 30 ]. WHO has also developed guidelines for GGT services related to these principles [ 11 ], which were also used in this work to adapt bioethical principles to GGT.
Accordingly, ETHICAL criteria were defined as those based on the bioethical principles of beneficence, non-maleficence and autonomy [ 29 ], including those criteria that refer to respect for human rights and dignity. Within the LEGAL field we considered criteria that provide guidelines to regulate the activities of the parties involved in GGT, particularly those that entail authority, limits and procedures for decision-making—what is decided, who decides, and how is decided—, so as to guarantee rights protection of those involved. Finally, SOCIAL criteria were defined as those referring to the principle of justice, understood as distributive justice based on what would be desirable to achieve in an equitably just society. Therefore, we have included criteria focused on activities that allow access to genomic medicine services and communication, as well as dissemination of information to different spheres of society.
Thus, whole scheme with each field that makes up ELSI and the connections between fields, is illustrated synoptically in Fig. 2 .
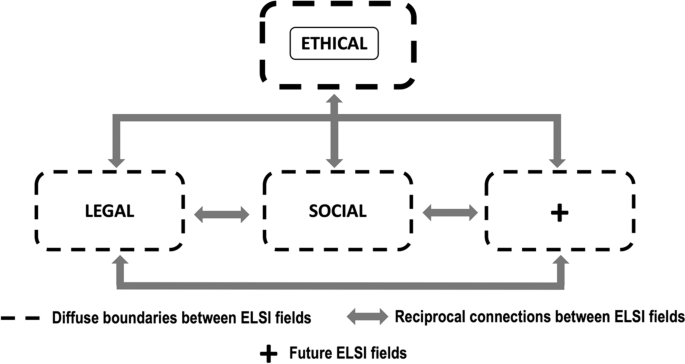
ELSI concept fields and their interconnections. The ETHICAL field is located at the top as mainstay for the rest of the fields—LEGAL, SOCIAL and future (+)—, which are placed in a lower hierarchical order. The interconnections between fields and the fuzzy limits among them are also represented
In conceptualizing the definitions of ELSI fields to build Fig. 2 , we realized the ETHICAL field encompasses respect for human rights and prevails over all scientific or economic interests, acting as the pillar for the rest of the fields since it connects vertically and transversally with all of them; therefore, it is placed at the head of the scheme and enclosed within a continuous line box. After all, the main topic on GGT testing are based on universal principles inherent to the patient as a human being. Subsequently, we placed in a lower hierarchical order the legal and social fields, plus an unnamed field labeled with the " + " sign in anticipation of future inclusion of additional fields to the ELSI studies. All fields are delimited by dotted-line boxes to indicate fuzzy boundaries between them, as it is common for their topics of interest to overlap. Bidirectional arrows indicate reciprocal connections between all fields since there are always relationships between them, for a particular criterion usually covers two or more fields and can hardly be studied in isolation. As noted in Fig. 2 there is substantial synergy between all components of ELSI, though it was possible to disintegrate the whole scheme into its individual parts—ETHICAL, LEGAL and SOCIAL fields. This configuration allows giving specific content to each of them and setting their respective parameters, while permitting addition of future elements in an orderly manner, so it helped to proceed with the rest of the study in an easier and more organized way.
Results and discussion
Elsi criteria on genetic-genomic testing in bioethical international documents.
A first general screening for the last 30-years period yielded a total of forty-four relevant sources, which once screened by recent publication dates and content close to GGT in terms of ethical, legal and social issues allowed us a final selection of nine documents for further detailed analysis (Table 1 ).
Other documents served as precedents since they also address bioethical issues related to human genetics, such as Nuremberg Code (1947) [ 31 , 32 ], Helsinki Declaration (1964) [ 33 ], Asilomar Conference on the Risks of Recombinant DNA (1975) [ 34 ], Belmont Report (1978) [ 35 ], Declaration of Lisbon on the Rights of the Patient (1981) [ 36 ], Report of the Nuffield Council on the ethical issues of genetic screening (1993) [ 37 ], Genomics and World Health (2002) [ 19 ], Declaration on Bioethics and Human Rights (2005) [ 38 ], among others. These documents are important in their own fields and historical moments but were not included in this study because they are already considered in the main updated texts finally selected. Although these international documents include other branches of genomic medicine—like research, cloning, genetic editing, etc.—, we focus on the closest for addressing specifically the ELSI aspects of GGT.
We identified documents of two types, international instruments and international recommendations. International non-binding instruments, like UDHG (General Conference, 29th, 1997) and HGD (General Conference, 32nd, 2003), were approved by a majority at general conferences of UNESCO and therefore are not signed by member countries. For OC and APOC, they become binding instruments for signatories’ countries. Recommendations documents were prepared by international panels of experts and coordinated by the international organizations (documents REI, DR, MGS, GQA, IBC) thus they are not signed or approved by countries. Most selected documents show a supreme interest in the prevalence of ethical aspects, i.e., safeguarding human rights and dignity above any other economic, social, commercial or research interests (as we will demonstrate below in a further analysis). And, although they share specific criteria in relation to ELSI issues, there are differences on which of these should apply to GGT, as well as in their depth. The following particularities merit special attention in chronological order:
The UNESCO 1997 Universal Declaration on the Human Genome and Human Rights (UDHG) [ 39 ] is considered the cornerstone in the international legal framework of ethical principles for genomic medicine and has served as the basis for all subsequent documents on this matter. It emphasizes the supreme interest of human rights, dignity and fundamental freedom over any other interest.
The Oviedo Convention (OC) [ 40 ] held that same year is the first internationally binding document for the countries that sign up and ratify it. To date, only 29 countries have done so [ 41 ]. It expands the principles of the UDGH on which it is based, specifying criteria on research topics, “Informed consent,” and “Genetic counseling”, also promoting the use of genetic tests exclusively for health purposes.
The Review of Ethical Aspects in Genetic Medicine (REI) [ 11 ], published by WHO in 2003, is based—like the present study—on the four principles of bioethics proposed by Beauchamp and Childress in 1979 (autonomy, beneficence, non-maleficence and justice). It takes these into the context of the ethical principles involved in genetic health care services for different patient groups, and of the technological applications it addresses. Focusing on the medical provider-patient relationship, it delves into the desired characteristics of informed consent and advice from health personnel to the patient. This document sets public education as a critical factor in the development of genetic services, considering social factors such as the response of diagnosed people and the social attitudes in different groups, with descriptions of advantages, risks and circumstances recommended to perform GGT.
The International Declaration on Human Genetic Data (HGD) [ 42 ], published by UNESCO in 2003, establishes the principles for collecting, processing, using and storing human genetic and protein data, and the biological samples from which such data originate. Although it is aimed at medical and scientific research, its principles are extended also to other service areas of genomic medicine, including cross-data topics as well as their eventual destruction of biological samples, physical and electronical records of human genetic data. In addition, it establishes the need to create regulations and urges countries to work on them for regulating cross-border transfer of human genetic data, proteomic data and biological samples, in order to promote international medical and scientific cooperation and guarantee equitable access to such data.
The Declaration of Reykjavik (DR) on Genetics and Medicine [ 12 ], published by the World Medical Association originally in 2005 and updated by the end of 2019, covers the ethical aspects of medical practice in research and clinical practice. It approaches GGT from the perspective of responsibility of physicians in the previous and later stages of interpreting results, including a section on unexpected findings, elaborating on the contents of genetic counseling with details on the characteristics that preparation of health care professionals should comprise to assure a broad informed consent.
The 2006 WHO report on Medical Genetic Services in Developing Countries (MGS) [ 8 ] covers the ethical, legal and social implications of genetic testing and screening. It is a comprehensive document with a greater focus on the subjects of interest. It highlights the principle of distributive justice and includes social issues which block genetic medicine services in developing countries. It recognizes the importance of protecting the privacy and confidentiality of genetic data to avoid discrimination and stigmatization in society, underlying the importance and priority of education and open dialogue about genetic medicine for the benefit of both society and the patient. Hence, it encourages actions that facilitate decision-making, such as counseling and the creation of patient-support organizations. It further analyzes the safety and well-being of the patient through quality assurance in products and services, particularly by strengthening regulations on those related to genetic matters. Finally, recommendations are issued to improve the ELSI criteria for genomic medicine in developing countries.
The Guidelines for Quality Assurance in Molecular Genetic Testing (GQA) [ 43 ], published by the OECD in 2007, promote a minimum of international standards for ensuring the quality of practices in molecular genetic testing laboratories, through compliance with the international quality standards ISO 17025 [ 44 ] for laboratory accreditation, testing and calibration, as well as with the ISO 15189 [ 45 ] standards for medical laboratories. In addition, this document encourages international cooperation and increases confidence of society in the governance of molecular genetic testing, laboratory surveillance, traceability of results, quality in reporting of results, and addresses the issue of cross-border exchange of samples and information.
The 2008 Additional Protocol to the Oviedo Convention on Genetic Testing for Health Purposes (APOC) [ 22 ] delves deeper than the original into the ethical and social principles of genetic testing. It provides more detail on sample types, communication of risk to family members, non-directive genetic counseling, quality of services, consent for people who cannot do it themselves, and respect for the user's private life. Currently, only six countries have signed and ratified this additional protocol [ 46 ].
The most comprehensive publication so far is that of UNESCO International Bioethics Committee of 2015 (IBC). In its Reflections on Human Genome and Human Rights [ 21 ], the document identifies five main ethical principles and social challenges: (a) respect for autonomy and privacy; (b) justice and solidarity; (c) understanding of health and disease; (d) cultural, social and economic context of science; and (e) responsibility for future generations. It exposes select topics of recently developed applications of genomic medicine, such as direct-to-consumer testing and personalized and precision medicine, assigning responsibility to the parties involved: countries, researchers, academics, physicians, regulators, for-profit companies and media, addressing also concerns about distributive justice and international solidarity. This is the more informative document presently available on the bioethics of genomic medicine with an interdisciplinary outlook.
The nine documents above vary in terms of their overall perspectives and approaches, depending on the respective publishing organization or entity, but a few main themes are prevalent in some of them: (a) clinical care and doctor-patient relationship (OC, REI, DR); (b) protection of patient rights through quality services (HGD, GQA); (c) review of interdisciplinary ELSI issues (MGS, IBC). The UDHG and the OC published in 1997 responded to international concerns about the applications of scientific and technological advances in relation to human health. Both documents focus on the protection of human rights and dignity during clinical research, so they concentrate on the ETHICAL field of ELSI and only superficially touch on the SOCIAL and LEGAL fields. In contrast, the 2015 IBC document deals mostly on ELSI criteria for GGT, going beyond the ethical aspect of rights protection and into the legal and social aspects of the commercialization of GGT technologies. It covers the protection of rights for patients as required by the ethical aspect but proceeds to assessing explicit legal responsibilities by each of the participants. In exploring the social aspect, IBC encompasses activities to ensure vertical and horizontal flow of information about GGT, and the distributive justice condition of securing access to services and international collaboration. It is clear, therefore, that progressive development has occurred from the first documents in the late 1990s, concerned mainly with ethical aspects for protection of rights for patients in research involving human beings, to more recent documents where the topics of technology applications and commercialization of products and services have become steadily included from about 2020.
This same progressiveness, moving from the protection of human rights in research to concerns about the use and commercialization of technology, is observed with the emergence of transnational ELSI programs. The first programs to study ELSI aspects of the human genome, such as the National Human Genome Research Institute of the United States in 1990 [ 47 ], pursued ethical and legal aspects focused on research. In turn, the programs emerged later were directed to multidisciplinary topics such as (a) programs focused on supporting public policy development and decision making: the "GE3LS" program of the Government of Canada in 2000 [ 48 ], the “p3G2" program of McGill University in 2004 [ 49 ]; (b) oriented to international scientific cooperation for research in genomics and society: the program "ELSI2.0" in 2012 [ 50 ]; and finally (c) focused on data sharing: the program "GA4GH" in 2013 [ 51 ]. All of these programs denote the change in interest about human genome studies, from just individual and family rights protection up to exploring many other issues in technology application to human health, stressing the need of creating regulations that guarantee the protection of genetic data and promote international medical and scientific cooperation.
Since no document covers all ELSI criteria by itself, nor they provide similar coverage of GGT, it seemed appropriate to select several documents and analyze them in depth so as to appreciate the fullness of the ELSI criteria related to GGT, for finally integrating these into a single extract as follows below.
Ethical, legal, and social criteria for genetic/genomic testing
As shown in Table 2 , 29 ELSI sub-criteria related to GGT were identified, which could be then organized and grouped into ten basic criteria: two in the ETHICAL field, four in the LEGAL field, and four in the SOCIAL field.
The ETHICAL field was confirmed as the support for all the analyzed documents, since all three present fields note the importance of protecting human rights and dignity as prime ethical criteria, which in turn affect the legal and social spheres. Here are found criteria such as "Patient Rights" and "Non-discrimination," which correspond to the bioethical principles of beneficence, autonomy, and non-maleficence. The "Patient Rights" criterion groups together those related to undergoing a GGT test and are valid from the moment the patient arrives at a health institute, whether public or private, until the results are safeguarded indefinitely in the files of the responsible laboratories or destroyed, depending on the specific situation. The criterion of "Non-discrimination" refers to the patients right to be treated equally inside and outside the health institution, regardless of the results of their genetic/genomic analyses as stated in the Report of the International Bioethics Committee on the Principle of Non-discrimination and Non-stigmatization [ 52 ]. It comprises three sub-criteria: (a) “Avoidance of reductionism” on the overestimation of genetic influence and underestimation of behavioral, psychosocial and environmental factors; (b) “Genetic exceptionalism” refers to the special handling of genetic data given its nature of providing information on the current or future state of health the person, their family and may also have cultural significance; (c) “Avoid stigmatization” of a person because of a test result for him or her, the family, group or community. The “Non-discrimination” criterion is recognized as a fundamental right of patients and is the most referenced in the documents here analyzed, so we divided it into three sub-criteria (Table 2 ). The ethical aspect is present in all the analyzed documents, since the ELSI studies began by covering the ethical field and therefore it has been examined for a longer time. Both "Patient Rights" and "Non-discrimination" are criteria that must always be accomplished and include mechanisms to be put into effect throughout the testing process. They represent a cross-cutting field through all other aspects of ELSI.
In fact, an analysis of each of nine documents that denotes the number of ethical, legal, or social sub-criteria covered and the respective percentage for field, showed that most of documents are oriented into the ethical field (Fig. 3 ). Thus, the highest percentages were observed in following documents: UDHG 100%, OC 75%, HGD 88%, DR 63%, APOC 88% and MGS 63% (which practically tied to the social field, 64%). While REI and IBC documents have a greater number of criteria covered in the social field, and only GQA stands out in the legal field, of course related to quality and regulation issues. These data support the assumption that some criteria might be incomplete in a single document and therefore the need to complement each other to build a more complete guideline.
Number of sub-criteria covered by each international document divided into ETHICAL, LEGAL and SOCIAL fields. The percentages indicate how much of the total identified sub-criteria is covered by each field
The LEGAL field contains most of the criteria and sub-criteria (fourteen) involved in GGT (Table 2 ), but they are not equally represented among the documents. Some of these, like the UDHG and OC, include only few legal sub-criteria like "Testing" and "Health Regulation"; furthermore, these documents do not delve into them. The "Protection of the Information" criterion focuses on time and form mechanisms to ensure safeguarding of biological samples, as well as any physical or electronic access to genetic information of the patient by unauthorized third parties, including destruction of both samples and data. The "Testing" criterion specifies under what circumstances it is advisable to perform a GGT test on the patient or a relative, informing the scope and limitations of the test. The "Health Regulation" criterion is the most extensive in content. It groups together the operations in which the legal regulatory and justice apparatus of each country must intervene to ensure the quality and reliability of GGT tests. The "Commercialization" criterion widens this scope by including sub-criteria related to the provision of GGT services and products, up to integrating technology commercialization with ELSI considerations, as it is found in the GQA and IBC documents. Yet this criterion was the least approached, denoting the research focus of most of the documents and reflecting also the international scene, where there is still no total convergence in a specific regulatory legal path for GGT, a pending situation for which the present study might be beneficial. It will be up to each country adopting proper regulations according to its particular circumstances, considering the international standards mentioned in Table 2 and always taking into account the cross-cutting ethical aspects.
As regards the SOCIAL field, we found criteria that refer to the bioethical principle of distributive justice, as well as criteria for accessing and communicating genetic information. Thus, four criteria are shown in Table 2 . "Genetic Counseling" describes the characteristics of such advice—non-directive, complete, with simple language, respectful—, which health professionals must carry out at the different stages of testing until solving all the doubts the patients might have. The "Training" criterion includes public policies in genomics, training of qualified human resources in this area, dissemination of related activities to different spheres of society, wide availability and accessibility of information on genetic testing through country institutions, as well as by academic and civil organizations. The "Reporting of Results" criterion as part of the social field, covers processes by which healthcare professionals communicate test results, associated information and address unexpected findings to the patients and in some cases to their families, this helps to mark the boundary to avoid genetic determinism. This means avoiding the consideration of a GGT result as a disease when this has not appeared, and ruling out other environmental, biological or psychosocial factors. Finally, the "Accessibility" criterion is intended to highlight the vital importance of finding ways by which GGT tests are made available to the entire population that could require them for health purposes, and not exclusively to those who can afford them. In fact, GGT accompanied by adequate medical counseling can serve as a routine instrument of public health care for opportune disease detection and prevention, and for reducing social inequality in this regard. Therefore, the "Genetic Counseling" criterion in its different phases of testing is the one that stands out the most. The other three SOCIAL criteria are well represented in the documents because of their importance, although general implementation guidelines for putting all of them into effect in countries are still lacking.
Priority criteria for genetic/genomic testing
It is also of vital importance to determine the priority criteria for genetic testing. Despite the relevance of each criterion here identified for GGT, not all of them are perceived with the same level of priority by experts in the international community. Criteria prioritization in the documents can be divided into four different groups. (A) Criteria that prioritize the safeguarding of dignity and human rights, (B) priority criteria to provide quality services regarding the protection of the health and the best interests for the patient, (C) prioritization approach to promote fair access to technology and health, (D) prioritization approach of the doctor-patient relationship.
As mentioned above, the scrutiny of the nine documents here analyzed shows differences in the attention given to the criteria and sub-criteria they contemplate. Figure 4 presents the coverage of the 29 sub-criteria ordered by numbers of citations in the analyzed documents.
GGT Sub-criteria arranged according to the number of documents in which they appear
The numbers of citations for sub-criteria may be divided into three groups (Fig. 4 ). The top group encompasses seven to nine citations (sub-criteria 1 to 8), including almost entirely those comprised in the two ETHICAL field criteria of "Patient Rights" and "Non-discrimination". This is expected, as the rights of patients constitute the basis for all ELSI documents referring to the human genome. Only one sub-criterion in this group belongs to the LEGAL field, for it concerns when it is valid applying a GGT test. This sub-criterion is highly referenced since these tests are recommended only for health purposes and under medical recommendation, seeking to discourage excessive use of GGT technology for mere curiosity (ancestry) or without proper medical indication, which could lead patients to making harmful decisions that might affect their health (direct-to-consumer tests or DTCs). As for criteria in the SOCIAL field, the top group includes "Genetic Counseling" due to the importance of medical monitoring in the diagnosis and treatment of genetic diseases. This top group includes sub-criteria with an A and D prioritization approach.
The middle group, with four to six citations, involves fourteen Sub-criteria (numbers 9 to 22), from which three belong to the LEGAL field—"Protection of the Information," "Testing," and especially the "Health Regulation"—, while other three correspond to the SOCIAL field—"Training," "Reporting of Results" and "Accessibility.” Therefore, middle group sub-criteria include B, C and D prioritization. The eight sub-criteria in the bottom group (numbers 23 to 29), turn up only between one and three times and mainly regarding the LEGAL criterion of "Commercialization" because, as pointed out previously, few documents contemplate the use and commercialization of GGT technology within ELSI studies. However, the most up-to-date review documents (MGS, GQA, IBC) have already incorporated this criterion. This bottom group is related to prioritization approach B.
In addition, Table 3 shows the criteria that we found relevant for a general prioritization approach in the three ELSI fields, corresponding to the urgency remarks pointed out by the analyzed documents from international organizations.
The seven priority criteria here distinguished are vastly represented in the analyzed documents, as shown in Fig. 4 . The ETHICAL priority criteria of "Patient Rights" and "Non-discrimination” (prioritization approach A), as well as the SOCIAL priority criterion of "Counseling" (prioritization approach D), appear at the top group in number of citations, and the rest of priority criteria in Table 3 occur in more than half of the documents analyzed. Moreover, the first two of such criteria, along with "Protection of the Information", have remained present from the first published documents elaborated with a purely ethical approach and their prioritization is still maintained. The "Health Regulation" criterion (prioritization approach B) was recognized as a priority about a decade after the first documents appeared, due to the growth in the use of GGT technology either for health purposes or other interests. The priorities of ensuring tests safety and the overall quality that laboratories provide to users are now widely acknowledged as well, as it occurs increasingly also with the “Accessibility” (prioritization approach C) and the “Training” criteria (prioritization approach B). We noticed progressivity in appreciation of ELSI criteria priorities, which is understandable, as their development over time responds to varying social, cultural and economic circumstances in different countries.
Our present observations for GGT are consistent with the WHO 2002 [ 19 ] deliberations on the ETHICAL and LEGAL priority criteria and the "Counseling" criterion. They pay particular attention to "Informed Consent" and protection of "Privacy and Confidentiality" to avoid discrimination. Furthermore, Granados-Moreno et al. [ 10 ] identified as especially relevant criteria for GGT "Counseling," "Validity and Clinical utility," "Confidentiality of information" and "Informed Consent", all of which are contained in and in agreement with our results. They considered also as relevant "Genetic Exceptionalism" and "Commercialization", including "Intellectual Property" and "Direct-to-consumer Testing", which seems to us a reasonable bioeconomy prospect for the rapid development of personalized medicine. WHO 2002 and Granados-Moreno et al. 2018 also identified as priority the "Reporting of results" criterion—"Communication of risks" and "Unexpected findings."—(prioritization approach D), and we certainly realize that the "Reporting of results" criterion is essential, since it appears in more than half of the documents here analyzed (Fig. 4 ). The prioritization approaches A, B and C, and even “Counseling” (prioritization approach D) can be supported by national legislations through laws, regulations and sanctions. Because our priority criteria are here aimed at primary wide-coverage efforts to define national GGT regulations beyond personalized GGT care, the “Reporting of results” criterion was not contemplated. Hence, it remains pending to seek synergy between health authorities, medical academies, physicians and patients to issue guidelines for the communication of results and their implications between doctor-patient, such as in the recent Declaration of Cordoba [ 53 ] and the Report of the International Bioethics Committee on the Principle of Individual Responsibility as related to Health [ 54 ].
The ELSI criteria have been progressing in step with GGT technological advancement and growing lobbying space for introducing health national laws. Although the UDHG is not an internationally binding document, several countries worldwide use it as a basis for developing their national laws. It certainly represents a good starting point; however, as already noted above, it was originally conceived to cover ethical issues in human genome research. Leaving aside other relevant and more complex aspects of ELSI studies—such as the use and commercialization of technology, the regulation of which is becoming increasingly necessary—, we firmly believe that widely accepted ELSI criteria, consistent with international bioethical requirements like those identified in the present work, could serve as a robust foundation for basing national regulations on personalized genomic medicine.
There are differences in the explicit content with which the ELSI criteria are addressed in the documents here analyzed—some of them, for example, offer an incomplete description by taking only a single document as reference—, and they would be more clearly understood if updated so as to complement each other. To do this, we propose the following: for the ETHICAL field, within the "Informed Consent" considerations, here limited to adults with full consent capacity (Table 2 ), specifications for people who cannot consent (by partial or null capacity to consent) to a GGT tests were also included under the OC (article 6 to 9), and expand according to APOC guidelines to obtain authorization (article 10 to 12), as well as to REI recommendations on autonomy and informed consent (see Table 5 of that document). For the LEGAL aspect: we suggest considering HGD guidelines on the collection, processing, use, and storage of genetic data and biological samples, both in research and by testing companies, including cross-border operations. As regards ensuring the quality of GGT services, we recommend supplementing the GQA principles and best practices (part I, section 2) with those of APOC on quality of genetic services and clinical utility (article 5 and 6), and IBC recommendations, particularly for DTC (issue 121). We perceived an urgency for GGT technology regulation, which would benefit from the inclusion of parameters on the attribution of responsibilities to the different stakeholders (States and governments, scientists and regulatory bodies, media and educators, economic actors and for-profit companies) as mentioned by IBC (page 3 and 4), and the coverage of damage repair pointed out in UDHG (article 8). The most comprehensive recommended reasons to applying GGT tests are found in APOC for benefit of patients or family members or for biological materials and deceased persons (articles 8, 13, 14 and 15), and may be supported by those found in IBC on the understanding of illness and health (section II.1.3). For the SOCIAL aspect: we propound taking the broad criteria of genetic counseling from REI about counseling competent adults, children and adolescents, persons with diminished mental capacity, adults who abdicate moral autonomy (part II section 2); in conjunction with DR recommendations for medical students and physicians giving counseling (page 2 and 3). To address unexpected findings, we found appropriate the guidelines presented in DR due to the explicit information that the patient should receive (page 2). To support the distributive justice, the recommendations for encouraging the formation of civilian organizations might be considered according to MGS (section 4.6.5) and REI (part I 3.5). Likewise, there is an intense call to promote bioethics education of both patients and society, according to the observations described in MGS for supporting the effectiveness of genetic services and help to prevent discrimination and stigmatization (section. 4.7); along with the vision of REI on education as the key to ethical genetics services (part I section 3), and finally with GQA principles for education and training standards for laboratory personnel (part I 2.E).
Synopsis and Perspectives
As shown above, the ethical, legal and social implications of research in the human genome constitute today a dynamic, progressive and profusely interconnected area of study. This is reflected in the evolution of published international documents and the parallel emergence of national ELSI programs. Three main attention areas stand out: (a) clinical care and doctor-patient relationship, (b) protection of rights for patients and relatives, and (c) advantages of interdisciplinary approach. Together they contribute to strengthen respect for human dignity, rights and privacy, in order to avoid discrimination for genetic reasons. The weakest points for effectively enforcing these demands lie in national health regulations and, above all, those related to commercialization of GGT technology.
As a necessary pending step to meet such challenges, here we present a first unified extract of the parameters that define each ELSI field and thus give actual meaning to the ethical, legal, and social issues in the major documents published on these subjects by international organizations in the last 30 years. Future elements can now be added to this disambiguated matrix in an orderly and organized manner, as needed to keep abreast with further developments in each field of study. In its present version it comprises the three fundamental ELSI fields relevant for genomic medicine, already customized for GGT: (1) applicable bioethical principles, (2) legal aspects in a flexible framework to fit the evolution of new technologies, and (3) the socio-cultural context of science and technology. It includes twenty-nine ELSI sub-criteria pertinent to GGT grouped into ten main criteria, along with seven top priority criteria we found to provide a precise starting point for reviewing, evaluating or proposing national regulations for GGT, in an integrative way for human rights protection and the development of bioeconomy.
The minimum criteria here identified are focused on GGT for medical purposes, but they can be easily extrapolated to other genomic medicine areas like biobanks [ 55 ], epigenetics [ 56 ], human genetic databases [ 57 ] and germline genetic modification [ 58 ]. It will undoubtedly be of interest extending this study to genetic testing of newborns and for non-medical purposes such as exploration of personal ancestry, as well as to groups of people in situations of vulnerability and with only partial capacity of consent like prisoners and mentally impaired persons. In addition, there are also criteria that become relevant in the international relations and research contexts, like "Genomic Sovereignty" [ 59 , 60 ], "Data Sharing" [ 61 , 62 ] and "International Cooperation" [ 63 , 64 ].
Given the multidisciplinary and dynamic nature of the ELSI fields, the incorporation of other complementary fields of study is to be expected. The Organization for Economic Co-operation and Development (OECD) recognizes that the main health challenges which countries and societies will face over the coming decades can be alleviated through the invention, development and use of products and processes of biological materials, a collection of factors designated as Bioeconomy [ 65 ]. And this assortment will certainly include legal and commercial aspects such as proof of innovation, registration of intellectual property, verification of functionality and safety, and authorization for sale and distribution, all of which go beyond issues relating directly to patient rights and health professional obligations. A practical example of the involvement of bioeconomy in genomic studies for society is the GE3LS program by a Canadian government organization [ 48 ], which considers even environmental and economic issues in addition to the usual ethical, legal, and social aspects. Since all of this will likely play a key role in future ELSI studies, particularly for the commercialization of GGT technology, we suggest the incorporation of a BIOECONOMY field into studies concerning genomic medicine.
On the other hand, as ELSI studies evolution transits from concerns about human rights protection to concerns about profitable applications of technology in genomic medicine, the urgency of proper harmonized regulations is evident. The harmonized ELSI criteria are useful to create, verify, and supplement national regulations, which may balance the development of bioeconomy, social equity, but above all provide the maximum protection of human rights and dignity before any economic, legal, commercial, social and research interests. However, there is still a long way to go for bringing on and pairing ELSI discussions into the field of GGT technology commercialization. Accordingly, in parallel to GGT technology fast advance, there is a considerable and pressing need for international harmonization of ELSI criteria in both content and quantity. For it is inaccurate and confusing to continue calling “ELSI criteria” to just a few somewhat arbitrarily used ones when there are at least two dozen of proper options, which can be narrowed down to concise meanings as shown in this paper. Hence, it is crucial for international and interdisciplinary panels of experts to discuss and stress the use of correct ELSI nomenclature in all contexts, for which we hope the present contribution may prove useful.
Availability of data and materials
Not applicable.
Abbreviations
Additional protocol to the convention on human rights and biomedicine, concerning genetic testing for health purposes
Deoxyribonucleic acid
Ethical considerations regarding the use of genetics in health care (declaration of Reykjavik)
Direct-to-consumer genetic testing
Ethical, legal, and social implications
Global alliance for genomics and health
Genomics and its ethical, environmental, economic, legal, and social aspects
Genetic and genomic testing
Guidelines for quality assurance in molecular genetic testing
International declaration on human genetic data
Report of the international bioethics committee on updating its reflection on the human genome and human rights
Medical genetic services in developing countries. The ethical, legal, and social implications of genetic testing and screening
Convention for the protection of human rights and dignity of the human being regarding the application of biology and medicine: convention on human rights and biomedicine (Oviedo convention)
Organization for economic co-operation and development
Policy partnerships project for genomic governance
Review of ethical issues in medical genetics
Universal Declaration on the Human Genome and Human Rights
United Nations Educational, Scientific and Cultural Organization
World Health Organization
World Medical Association
Khoury MJ. Genetics and genomics in practice: the continuum from genetic disease to genetic information in health and disease. Genet Med. 2003;5(4):261–8. https://doi.org/10.1097/01.GIM.0000076977.90682.A5 .
Article Google Scholar
Burke W. Genetic testing. N Engl J Med. 2002;347(23):1867–75. https://doi.org/10.1056/NEJMoa012113 .
Health IoMURoTG-BRf. Generating evidence for genomic diagnostic test development: workshop summary. the national academies collection: reports funded by national institutes of health. Washington (DC) 2011.
Patch C, Sequeiros J, Cornel MC. Genetic horoscopes: is it all in the genes? Points for regulatory control of direct-to-consumer genetic testing. Eur J Hum Genet. 2009;17(7):857–9. https://doi.org/10.1038/ejhg.2008.246 .
McEwen JE, Boyer JT, Sun KY, Rothenberg KH, Lockhart NC, Guyer MS. The ethical, legal, and social implications program of the national human genome research institute: reflections on an ongoing experiment. Annu Rev Genomics Hum Genet. 2014;15:481–505. https://doi.org/10.1146/annurev-genom-090413-025327 .
Burke W, Appelbaum P, Dame L, Marshall P, Press N, Pyeritz R, et al. The translational potential of research on the ethical, legal, and social implications of genomics. Genet Med. 2015;17(1):12–20. https://doi.org/10.1038/gim.2014.74 .
McWalter K, Gaviglio A. Introduction to the special issue: public health genetics and genomics. J Genet Couns. 2015;24(3):375–80. https://doi.org/10.1007/s10897-015-9825-9 .
Ballantyne A, Goold I, Pearn A, Programme WHOHG. Medical genetic services in developing countries: the ethical, legal and social implications of genetic testing and screening. World Health Organization, Geneva; 2006.
Nambisan P. Chapter 7—Genetic Testing, Genetic Discrimination and Human Rights. In: Nambisan P, editor. An introduction to ethical, safety and intellectual property rights issues in biotechnology. Academic Press: Cambridge; 2017. p. 171–87.
Chapter Google Scholar
Granados-Moreno P, Noohi F, Joly Y. Ethics and genetics ☆ . Reference module in biomedical sciences. Amsterdam: Elsevier; 2018.
Google Scholar
Wertz DC, Fletcher GF, Berg K, Programme WHOHG. In: DC Wertz, JC Fletcher, K Berg (eds) Review of ethical issues in medical genetics: report of consultants to WHO. World Health Organization, Geneva; 2003.
World Medical Association. WMA Declaration of Reykjavik—Ethical Considerations Regarding the Use of Genetics in Health Care. 2005.
Zhong A, Darren B, Loiseau B, He LQB, Chang T, Hill J, et al. Ethical, social, and cultural issues related to clinical genetic testing and counseling in low- and middle-income countries: a systematic review. Genet Med. 2018. https://doi.org/10.1038/s41436-018-0090-9 .
Metternick-Jones SC, Lister KJ, Dawkins HJS, White CA, Weeramanthri TS. Review of current international decision-making processes for newborn screening: lessons for Australia. Front Public Health. 2015;3:214. https://doi.org/10.3389/fpubh.2015.00214 .
Mello MM, Wolf LE. The havasupai Indian tribe case—lessons for research involving stored biologic samples. New Engl J Med. 2010;363(3):204–7. https://doi.org/10.1056/NEJMp1005203 .
Horton R, Crawford G, Freeman L, Fenwick A, Lucassen A. Direct-to-consumer genetic testing with third party interpretation: beware of spurious results. Emerg Top Life Sci. 2019;3(5):669–74. https://doi.org/10.1042/ETLS20190059 .
Caulfield T, Chandrasekharan S, Joly Y, Cook-Deegan R. Harm, hype and evidence: ELSI research and policy guidance. Genome Med. 2013;5(3):21. https://doi.org/10.1186/gm425 .
Parens E, Appelbaum PS. On what we have learned and still need to learn about the psychosocial impacts of genetic testing. Hastings Cent Rep. 2019;49(Suppl 1):S2–9. https://doi.org/10.1002/hast.1011 .
World Health Organization. Advisory Committee on Health Research. Genomics and world health report of the Advisory Committee on Health Research. World Health Organization, Geneva; 2002.
Connell S. Bioethics: ELSI. eLS. Hoboken: Wiley; 2001.
International Bioethics Committee. Report of the IBC on updating its reflection on the Human Genome and Human Rights. UNESCO; 2015.
Council of Europe. Additional Protocol to the Convention on Human Rights and Biomedicine concerning Genetic Testing for Health Purposes. 2008.
Strech D, Sofaer N. How to write a systematic review of reasons. J Med Ethics. 2012;38(2):121–6. https://doi.org/10.1136/medethics-2011-100096 .
Boyle AE. Some reflections on the relationship of treaties and soft law. Int Compar Law Q. 1999;48(4):901–13. https://doi.org/10.1017/S0020589300063739 .
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. PLoS Med. 2021;18(3): e1003583. https://doi.org/10.1371/journal.pmed.1003583 .
ELSAGEN. Ethical, legal and social aspects of human genetic databases. A European Comparison. 2002. http://www.elsagen.net/project.html . Accessed 22 Sep 2021.
Chameau J-L, Ballhaus WF, Lin H, National Research Council (U.S.). Committee on Ethical and Societal Implications of Advances in Militarily Significant Technologies That are Rapidly Changing and Increasingly Globally Accessible., National Research Council (U.S.). Committee on Science Technology and Law., National Academy of Engineering. Center for Engineering Ethics and Society Advisory Group. Emerging and readily available technologies and national security: a framework for addressing ethical, legal, and societal issues. National Academies Press, Washington, DC; 2014.
Olejarczyk JP YM. Patient Rights And Ethics. StatPearls [Internet]. 2021. https://www.ncbi.nlm.nih.gov/books/NBK538279/ .
Beauchamp TL, Childress JF. Principles of biomedical ethics. New York: Oxford University Press; 1979.
Erdmann A, Rehmann-Sutter C, Bozzaro C. Patients’ and professionals’ views related to ethical issues in precision medicine: a mixed research synthesis. BMC Med Ethics. 2021;22(1):116. https://doi.org/10.1186/s12910-021-00682-8 .
Weindling P. The origins of informed consent: the International Scientific Commission on Medical War Crimes, and the Nuremburg code. Bull Hist Med. 2001;75(1):37–71. https://doi.org/10.1353/bhm.2001.0049 .
Dörner K, Ebbinghaus A, Linne K, Roth KH, Weindling P, Eltzschig J et al. The Nuremberg Medical Trial, 1946/47: transcripts, material of the prosecution and defense, related documents. English ed. ed.
World Medical Association. WMA Declaration of Helsinki – Ethical Principles for Medical Research Involving Human Subjects. 1964.
Berg P, Baltimore D, Brenner S, Roblin RO, Singer MF. Summary statement of the Asilomar conference on recombinant DNA molecules. Proc Natl Acad Sci. 1975;72(6):1981–4.
United States. National Commission for the Protection of Human Subjects of Biomedical and Behavioral Research. The Belmont report : ethical principles and guidelines for the protection of human subjects of research. DHEW Publication no (OS) 78-0012. Bethesda, Md. Washington: The Commission; for sale by the Supt. of Docs., U.S. Govt. Print. Off.; 1978.
World Medical Association. WMA Declaration of Lisbon on the Rights of the Patient. 1981.
Nuffield Council on Bioethics. Genetic Screening Ethical Issues. 1993.
United Nations Educational Scientific and Cultural Organization. Universal Declaration on Bioethics and Human Rights. 2005.
United Nations Educational Scientific and Cultural Organization. Universal Declaration on the Human Genome and Human Rights. 1997.
Council of Europe. Convention for the protection of Human Rights and Dignity of the Human Being with regard to the Application of Biology and Medicine: Convention on Human Rights and Biomedicine. 1997.
Council of Europe. Chart of signatures and ratifications of Treaty 164. https://www.coe.int/en/web/conventions/full-list/-/conventions/treaty/203?module=signatures-by-treaty&treatynum=164 . Accessed 28 Jul 2020.
United Nations Educational Scientific and Cultural Organization. International Declaration on Human Genetic Data. 2003.
Organization for Economic Co-Operation and Development. OECD Guidelines for Quality Assurance in Molecular Genetic Testing. 2007.
International Organization for Standardization. Testing and Calibration Laboratories. International Organization for Standardization; 2017.
International Organization for Standardization. Medical laboratories—Requirements for quality and competence. International Organization for Standardization; 2012.
Council of Europe. Chart of signatures and ratifications of Treaty 203. https://www.coe.int/en/web/conventions/full-list/-/conventions/treaty/203?module=signatures-by-treaty&treatynum=203 . Accessed 28 Jul 2020.
National Human Genome Research Institute. Ethical, Legal and Social Implications Research Program. National Human Genome Research Institute. https://www.genome.gov/Funded-Programs-Projects/ELSI-Research-Program-ethical-legal-social-implications . Accessed 22 Jul 2020.
Genome Canada. Genomics in Society / GE3LS. https://www.genomecanada.ca/en/programs/genomics-society-ge3ls . Accessed 3 Jul 2020.
Centre of Genomics and Policy. Policy Partnerships Project for Genomic Governance (p3G2). McGill University. https://p3g2.org/about-p3g2/ . Accessed 22 Jul 2020.
The Global Health Network. ELSI2.0. https://elsi2workspace.tghn.org/ . Accessed 22 Jun 2020.
Global Alliance for Genomics and Health. Global Alliance for Genomics and Health (GA4GH). 2013. https://www.ga4gh.org/about-us/ . Accessed 22 Jun 2020.
International Bioethics Committee. Report of the IBC on the Principle of Non-discrimination and Non-stigmatization. UNESCO; 2014.
World Medical Association. WMA Declaration of Cordoba on Patient-Physician Relationship. 2020.
International Bioethics Committee. Report of the IBC on the Principle of Individual Responsibility as related to Health. 2019.
Bledsoe MJ. Ethical Legal and Social Issues of Biobanking: Past, Present, and Future. Biopreserv Biobank. 2017;15(2):142–7. https://doi.org/10.1089/bio.2017.0030 .
Joly Y, So D, Saulnier K, Dyke SOM. Epigenetics ELSI: Darker Than You Think? Trends Genet. 2016;32(10):591–2. https://doi.org/10.1016/j.tig.2016.07.001 .
Árnason V. Introduction: some lessons of ELSAGEN. In: Árnason G, Häyry M, Chadwick R, Árnason V, editors. The ethics and governance of human genetic database: European perspectives. Cambridge: Cambridge University Press; 2007. p. 1–8.
Chan S, Donovan PJ, Douglas T, Gyngell C, Harris J, Lovell-Badge R, et al. Genome Editing Technologies and Human Germline Genetic Modification: The Hinxton Group Consensus Statement. Am J Bioeth. 2015;15(12):42–7. https://doi.org/10.1080/15265161.2015.1103814 .
Siqueiros-García J, Oliva-Sanchez P, Saruwatari-Zavala G. Genomic sovereignty or the enemy within. Acta Bioethica. 2013;19:269–73. https://doi.org/10.4067/S1726-569X2013000200011 .
Labuschaigne M, Pepper M. “A room of our own?” Legal lacunae regarding genomic sovereignty in South Africa. J Contemp Roman-Dutch Law. 2010;73:432–50.
Balaji D, Terry SF. Benefits and risks of sharing genomic information. Genet Test Mol Biomarkers. 2015;19(12):648–9. https://doi.org/10.1089/gtmb.2015.29008.sjt .
Sorani MD, Yue JK, Sharma S, Manley GT, Ferguson AR, Investigators TT. Genetic data sharing and privacy. Neuroinformatics. 2015;13(1):1–6. https://doi.org/10.1007/s12021-014-9248-z .
Yoshizawa G, Ho CW, Zhu W, Hu C, Syukriani Y, Lee I, et al. ELSI practices in genomic research in East Asia: implications for research collaboration and public participation. Genome Med. 2014;6(5):39. https://doi.org/10.1186/gm556 .
Knoppers BM, Abdul-Rahman MH, Bédard K. Genomic databases and international collaboration. King’s Law J. 2007;18(2):291–311. https://doi.org/10.1080/09615768.2007.11427678 .
Organisation for Economic Co-operation and Development, OECD International Futures Programme. The bioeconomy to 2030 designing a policy agenda. Organization for Economic Co-operation and Development, Paris; 2009.
Download references
Acknowledgements
The first author was supported by a doctoral scholarship (243314) from the Consejo Nacional de Ciencia y Tecnología (CONACyT México). We thank for Open Access funding provided by Cinvestav and Inmegen.
Author information
Authors and affiliations.
Program of Science, Technology and Society, Center for Research and Advanced Studies IPN (Cinvestav), 07360, Mexico City, Mexico
Tania Ascencio-Carbajal, Fernando Navarro-Garcia & Eugenio Frixione
Department of Cell Biology, Center for Research and Advanced Studies IPN (Cinvestav), 07360, Mexico City, Mexico
Fernando Navarro-Garcia & Eugenio Frixione
Department of Legal, Ethical and Social Studies, National Institute of Genomic Medicine (Inmegen), 14610, Mexico City, Mexico
Garbiñe Saruwatari-Zavala
You can also search for this author in PubMed Google Scholar
Contributions
TAC conceptualized the idea, searched for literature, analyzed the information, wrote first draft of the manuscript and edited all versions. GSZ contributed to conceptual work, analyzed results, provided bibliographic material and reviewed first versions of the manuscript. FNG and EF contributed to conceptual work, analyzed results, reviewed and edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Correspondence to Fernando Navarro-Garcia or Eugenio Frixione .
Ethics declarations
Ethics approval and consent to participate, consent for publication, competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Ascencio-Carbajal, T., Saruwatari-Zavala, G., Navarro-Garcia, F. et al. Genetic/genomic testing: defining the parameters for ethical, legal and social implications (ELSI). BMC Med Ethics 22 , 156 (2021). https://doi.org/10.1186/s12910-021-00720-5
Download citation
Received : 09 August 2021
Accepted : 19 October 2021
Published : 23 November 2021
DOI : https://doi.org/10.1186/s12910-021-00720-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Genetic testing
- Genomic testing
- Legal and social implications
- Genomic medicine
- ELSI criteria
- Genomic diseases
- Patient rights
- Policy-making
BMC Medical Ethics
ISSN: 1472-6939
- General enquiries: [email protected]
Research Topics
The Center for Genetic Medicine’s faculty members represent 33 departments or programs across three Northwestern University schools and three Feinberg-affiliated healthcare institutions. Faculty use genetics and molecular genetic approaches to understand biological processes for a diverse range of practical and clinical applications.
Select a topic below to learn more and see a list of faculty associated with that type of research. For a full list of Center for Genetic Medicine members, visit our Members section .
Animal Models of Human Disease
Using genetic approaches with model organisms to investigate cellular and physiological processes can lead to improved approaches for detection, prevention and treatment of human diseases.
VIEW MEMBERS
Bioinformatics & Statistics
Bioinformatics, a discipline that unites biology, computer science, statistical methods, and information technology, helps researchers understand how genes or parts of genes relate to other genes, and how genes interact to form networks. These studies provide insight to normal cellular functions and how these functions are disturbed by disease. Statistics is central to genetic approaches, providing quantitative support for biological observations, and statistical genetics is heavily used by laboratories performing gene and trait mapping, sequencing and genotyping, epidemiology, population genetics and risk analysis.
Cancer Genetics and Genomics
Cancer begins with genetic changes, or mutations, that disrupt normal regulation of cell proliferation, survival and death. Inherited genetic changes contribute to the most common cancers, like breast and colon cancer, and genetic testing can help identify risks for disease. Tumors also develop additional genetic changes, or somatic mutations, that promote cancer growth and tumor metastases. These genetic changes can be readily defined through DNA and RNA sequencing. Genetic changes within a tumor can be used to develop and guide treatment options.
Cardiovascular Genetics
Cardiovascular disease is one of the leading causes of death in the US, and the risk of cardiovascular disease is highly dependent inherited genetic changes. The most common forms of heart disease including heart failure, arrhythmias, and vascular disease are under heritable genetic changes. We work to identify and understand the functions of genes that affect the risk of developing cardiovascular disease, as well as to understand the function of genes involved in the normal and pathological development of the heart.
Clinical and Therapeutics
Using genetic data identifies pathways for developing new therapies and applying existing therapies. DNA sequencing and epigenetic profiling of tumors helps define the precise defects responsible for cancer progression. We use genetic signals to validate pathways for therapy development. We are using gene editing methods to correct genetic defects. These novel strategies are used to treat patients at Northwestern Memorial Hospital and the Ann & Robert H. Lurie Children's Hospital of Chicago.
Development
The genomic blueprint of a single fertilized egg directs the formation of the entire organism. To understand the cellular processes that allow cells to create organs and whole animals from this blueprint, we use genetic approaches to investigate the development of model organisms and humans. Induced pluripotent stem cells can be readily generated from skin, blood or urine cells and used to mirror human developmental processes. These studies help us define how genes coordinate normal human development and the changes that occur in diseases, with the goal of improving detection, prevention and treatment of human disease.
Epigenetics/Chromatin Structure/Gene Expression
Abnormal gene expression underlies many diseases, including cancer and cardiovascular diseases. We investigate how gene expression is regulated by chromatin structure and other regulators to understand abnormal gene expression in disease, and to learn how to manipulate gene expression for therapeutic purposes.
Gene Editing/Gene Therapy
Gene editing tools like CRISPR/Cas can be used to directly alter the DNA code. This tool is being used to generate cell and animal models of human diseases and disease processes. Gene therapy is being used to treat human disease conditions.
Genetic Counseling
As part of training in genetic counseling, each student completes a thesis project. These projects examine all aspects of genetic counseling ranging from family-based studies to mechanisms of genetic action. With the expansion of genetic testing, genetic counselors are now conducting research on outcomes, cost effectiveness, and quality improvement.
Genetic Determinants of Cellular Biology
Genetic mutations ultimately change the functionality of the cells in which they are found. Mutations in genes encoding nuclear, cytoplasmic and extracellular matrix protein lead to many different human diseases, ranging from neurological and developmental disorders to cancer and heart disease. Using induced pluripotent stem cell and gene-editing technologies, it is now possible to generate and study nearly every human genetic disorder. Having cellular models of disease is necessary to develop new treatments.
Immunology
Many immunological diseases, such as Rheumatoid arthritis, Lupus, scleroderma, and others have a genetic basis. We work to understand how genetic changes and misregulation contribute to immunological diseases, and use genetic approaches to investigate how the immune system functions.
Infectious Disease/Microbiome
The susceptibility and/or pathological consequences of many infectious diseases have a genetic basis. We investigate how human genes interact with infectious diseases, and use genetic approaches to determine the interactions between pathogens and the host. Genetic tools, including deep sequencing, are most commonly used to define the microbiome as it undergoes adaptation and maladaptation to its host environment.
Neuroscience
We work to understand how genes contribute to neurological diseases, and use genetic approaches to investigate how the nervous system functions. Epilepsy, movement disorders, and dementia are heritable and under genetic influence. Neuromuscular diseases including muscular dystrophies and myopathies arise from primary mutations and research in genetic correction is moving into human trials and drug approvals.
Population Genetics/Epidemiology
Genetic data is increasingly available from large human populations and is advancing the population-level understanding of genetic risk. Northwestern participates in All-Of-US, which aims to build a cohort of one million citizens to expand genetic knowledge of human diseases. Race and ancestry have genetic determinants and genetic polymorphisms can help mark disease risks better than other markers of race/ancestry. We use epidemiology and population genetics to investigate the genetic basis of disease, and to assess how genetic diseases affect subgroups within broader populations.
Reproduction
Research is examining how germ cells are specified. We study the broad range of biology required to transmit genetic information from one generation to another, and how to facilitate the process of reproduction when difficulties arise or to avoid passing on mutant genes.
Follow Center for Genetic Medicine on Twitter Flickr
- Write my thesis
- Thesis writers
- Buy thesis papers
- Bachelor thesis
- Master's thesis
- Thesis editing services
- Thesis proofreading services
- Buy a thesis online
- Write my dissertation
- Dissertation proposal help
- Pay for dissertation
- Custom dissertation
- Dissertation help online
- Buy dissertation online
- Cheap dissertation
- Dissertation editing services
- Write my research paper
- Buy research paper online
- Pay for research paper
- Research paper help
- Order research paper
- Custom research paper
- Cheap research paper
- Research papers for sale
- Thesis subjects
- How It Works
122 The Best Genetics Research Topics For Projects

The study of genetics takes place across different levels of the education system in academic facilities all around the world. It is an academic discipline that seeks to explain the mechanism of heredity and genes in living organisms. First discovered back in the 1850s, the study of genetics has come a pretty long way, and it plays such an immense role in our everyday lives. Therefore, when you are assigned a genetics research paper, you should pick a topic that is not only interesting to you but one that you understand well.
Choosing Research Topics in Genetics
Even for the most knowledgeable person in the room, choosing a genetics topic for research papers can be, at times, a hectic experience. So we put together a list of some of the most exciting top in genetics to make the endeavor easier for you. However, note, while all the topics we’ve listed below will enable you to write a unique genetic project, remember what you choose can make or break your paper. So again, select a topic that you are both interested and knowledgeable on, and that has plenty of research materials to use. Without further ado, check out the topics below.
Interesting Genetics Topics for your Next Research Paper
- Genes and DNA: write a beginners’ guide to genetics and its applications
- Factors that contribute or/and cause genetic mutations
- Genetics and obesity, what do you need to know?
- Describe RNA information
- Is there a possibility of the genetic code being confidential?
- Are there any living cells present in the gene?
- Cancer and genetics
- Describe the role of genetics in the fight against Alzheimer’s disease
- What is the gene
- Is there a link between genetics and Parkinson’s disease? Explain your answer.
- Replacement of genes and artificial chromosomes
- Explain genetic grounds for obesity
- Development and disease; how can genetics dissect the developing process
- Analyzing gene expression – RNA
- Gene interaction; eye development
- Advances and developments in nanotechnology to enable therapeutic methods for the treatment of HIV and AIDS.
- Isolating and identifying the cancer treatment activity of special organic metal compounds.
- Analyzing the characteristics in certain human genes that can withstand heavy metals.
- A detailed analysis of genotypes that is both sensitive and able to endure heavy metals.
- Isolating special growth-inducing bacteria that can assist crops during heavy metal damage and identifying lipid directing molecules for escalating heavy metal endurance in plants.
Hot and Controversial Topics in Genetics
- Is there a link between genetics and homosexuality? Explain your answer
- Is it ethical and morally upright to grow human organs
- Can DNA changes beat aging
- The history and development of human cloning science
- How addictive substances alter our genes
- Are genetically modified foods safe for human and animal consumption?
- Is depression a genetically based condition?
- Genetic diagnosis of the fetus
- Genetic analysis of the DNA structure
- What impact does cloning have on future generations?
- What is the link between genetics and autism?
- Can artificial insemination have any sort of genetic impact on a person?
- The advancements in genetic research and the bioethics that come with them.
- Is human organ farming a possibility today?
- Can genetics allow us to design and build a human to our specifications?
- Is it ethical to try and tamper with human genetics in any way?
Molecular Genetics Topics
- Molecular techniques: How to analyze DNA(including genomes), RNA as well as proteins
- Stem cells describe their potential and shortcomings
- Describe molecular and genome evolution
- Describe DNA as the agent of heredity
- Explain the power of targeted mutagenesis
- Bacteria as a genetic system
- Explain how genetic factors increase cancer susceptibility
- Outline and describe recent advances in molecular cancer genetics
- Does our DNA sequencing have space for more?
- Terminal illness and DNA.
- Does our DNA determine our body structure?
- What more can we possibly discover about DNA?
Genetic Engineering Topics
- Define gene editing, and outline key gene-editing technologies, explaining their impact on genetic engineering
- The essential role the human microbiome plays in preventing diseases
- The principles of genetic engineering
- Project on different types of cloning
- What is whole genome sequencing
- Explain existing studies on DNA-modified organisms
- How cloning can impact medicine
- Does our genetics hold the key to disease prevention?
- Can our genetics make us resistant to certain bacteria and viruses?
- Why our genetics plays a role in chronic degenerative diseases.
- Is it possible to create an organism in a controlled environment with genetic engineering?
- Would cloning lead to new advancements in genetic research?
- Is there a possibility to enhance human DNA?
- Why do we share DNA with so many other animals on the planet?
- Is our DNA still evolving or have reached our biological limit?
- Can human DNA be manipulated on a molecular or atomic level?
- Do we know everything there is to know about our DNA, or is there more?
Controversial Human Genetic Topics
- Who owns the rights to the human genome
- Is it legal for parents to order genetically perfect children
- is genetic testing necessary
- What is your stand on artificial insemination vs. ordinary pregnancy
- Do biotech companies have the right to patent human genes
- Define the scope of the accuracy of genetic testing
- Perks of human genetic engineering
- Write about gene replacement and its relationship to artificial chromosomes.
- Analyzing DNA and cloning
- DNA isolation and nanotechnology methods to achieve it.
- Genotyping of African citizens.
- Greatly mutating Y-STRs and the isolated study of their genetic variation.
- The analytical finding of indels and their genetic diversity.
DNA Research Paper Topics
The role and research of DNA are so impactful today that it has a significant effect on our daily lives today. From health care to medication and ethics, over the last few decades, our knowledge of DNA has experienced a lot of growth. A lot has been discovered from the research of DNA and genetics.
Therefore, writing a good research paper on DNA is quite the task today. Choosing the right topic can make things a lot easier and interesting for writing your paper. Also, make sure that you have reliable resources before you begin with your paper.
- Can we possibly identify and extract dinosaur DNA?
- Is the possibility of cloning just around the corner?
- Is there a connection between the way we behave and our genetic sequence?
- DNA research and the environment we live in.
- Does our DNA sequencing have something to do with our allergies?
- The connection between hereditary diseases and our DNA.
- The new perspectives and complications that DNA can give us.
- Is DNA the reason all don’t have similar looks?
- How complex human DNA is.
- Is there any sort of connection between our DNA and cancer susceptibility and resistance?
- What components of our DNA affect our decision-making and personality?
- Is it possible to create DNA from scratch under the right conditions?
- Why is carbon such a big factor in DNA composition?
- Why is RNA something to consider in viral research and its impact on human DNA?
- Can we detect defects in a person’s DNA before they are born?
Genetics Topics For Presentation
The subject of genetics can be quite broad and complex. However, choosing a topic that you are familiar with and is unique can be beneficial to your presentation. Genetics plays an important part in biology and has an effect on everyone, from our personal lives to our professional careers.
Below are some topics you can use to set up a great genetics presentation. It helps to pick a topic that you find engaging and have a good understanding of. This helps by making your presentation clear and concise.
- Can we create an artificial gene that’s made up of synthetic chromosomes?
- Is cloning the next step in genetic research and engineering?
- The complexity and significance of genetic mutation.
- The unlimited potential and advantages of human genetics.
- What can the analysis of an individual’s DNA tell us about their genetics?
- Is it necessary to conduct any form of genetic testing?
- Is it ethical to possibly own a patent to patent genes?
- How accurate are the results of a genetics test?
- Can hereditary conditions be isolated and eliminated with genetic research?
- Can genetically modified food have an impact on our genetics?
- Can genetics have a role to play in an individual’s sexuality?
- The advantages of further genetic research.
- The pros and cons of genetic engineering.
- The genetic impact of terminal and neurological diseases.
Biotechnology Topics For Research Papers
As we all know, the combination of biology and technology is a great subject. Biotechnology still offers many opportunities for eager minds to make innovations. Biotechnology has a significant role in the development of modern technology.
Below you can find some interesting topics to use in your next biotechnology research paper. Make sure that your sources are reliable and engage both you and the reader.
- Settlements that promote sustainable energy technology maintenance.
- Producing ethanol through molasses emission treatment.
- Evapotranspiration and its different processes.
- Circular biotechnology and its widespread framework.
- Understanding the genes responsible for flora response to harsh conditions.
- Molecule signaling in plants responding to dehydration and increased sodium.
- The genetic improvement of plant capabilities in major crop yielding.
- Pharmacogenomics on cancer treatment medication.
- Pharmacogenomics on hypertension treating medication.
- The uses of nanotechnology in genotyping.
- How we can quickly detect and identify food-connected pathogens using molecular-based technology.
- The impact of processing technology both new and traditional on bacteria cultures linked to Aspalathus linearis.
- A detailed analysis of adequate and renewable sorghum sources for bioethanol manufacturing in South Africa.
- A detailed analysis of cancer treatment agents represented as special quinone compounds.
- Understanding the targeted administering of embelin to cancerous cells.
Tips for Writing an Interesting Genetics Research Paper
All the genetics research topics above are excellent, and if utilized well, could help you come up with a killer research paper. However, a good genetics research paper goes beyond the topic. Therefore, besides choosing a topic, you are most interested in, and one with sufficient research materials ensure you
Fully Understand the Research Paper Format
You may write on the most interesting genetics topics and have a well-thought-out set of ideas, but if your work is not arranged in an engaging and readable manner, your professor is likely to dismiss it, without looking at what you’ve written. That is the last thing you need as a person seeking to score excellent grades. Therefore, before you even put pen to paper, understand what research format is required.
Keep in mind that part of understanding the paper’s format is knowing what words to use and not to use. You can contact our trustful masters to get qualified assistance.
Research Thoroughly and Create an Outline
Whichever genetics research paper topics you decide to go with, the key to having excellent results is appropriately researching it. Therefore, embark on a journey to understand your genetics research paper topic by thoroughly studying it using resources from your school’s library and the internet.
Ensure you create an outline so that you can note all the useful genetic project ideas down. A research paper outline will help ensure that you don’t forget even one important point. It also enables you to organize your thoughts. That way, writing them down in the actual genetics research paper becomes smooth sailing. In other words, a genetics project outline is more like a sketch of the paper.
Other than the outline, it pays to have an excellent research strategy. In other words, instead of looking for information on any random source you come across, it would be wise to have a step-by-step process of looking for the research information.
For instance, you could start by reading your notes to see what they have to say about the topic you’ve chosen. Next, visit your school’s library, go through any books related to your genetics research paper topic to see whether the information on your notes is correct and for additional information on the topic. Note, you can visit the library either physically or via your school’s website. Lastly, browse educational sites such as Google Scholar, for additional information. This way, you’ll start your work with a bunch of excellent genetics project ideas, and at the same time, you’ll have enjoyed every step of the research process.
Get Down to Work
Now turn the genetics project ideas on your outline into a genetics research paper full of useful and factual information.
There is no denying writing a genetics research paper is one of the hardest parts of your studies. But with the above genetics topics and writing tips to guide you, it should be a tad easier. Good luck!
Leave a Reply Cancel reply
Genetics Topics for Presentation: 100+ Comprehensive Ideas
Explore 100+ detailed genetics presentation topics, from basic concepts to advanced research, genetic disorders, and ethical considerations. Perfect for any audience.
Understanding Genetics: An Overview
Categories of genetics topics.
- Fundamentals of Genetics
- Genetic Disorders
- Molecular Genetics
- Population Genetics
- Genetics in Medicine
- Genetic Engineering and Biotechnology
- Ethics in Genetics
- Evolutionary Genetics
- Genetics and Society
- Emerging Trends in Genetics
1. Fundamentals of Genetics
- Mendelian Genetics: Understanding Gregor Mendel’s laws of inheritance.
- DNA Structure and Function: The blueprint of life.
- Gene Expression and Regulation: How genes turn on and off.
- Chromosomes and Chromosomal Disorders: Understanding the role of chromosomes in genetics.
- Genetic Mutations: Types, causes, and effects.
- The Human Genome Project: Mapping human genes.
- Genotype vs. Phenotype: What you inherit vs. what you see.
- Epigenetics: How lifestyle and environment can affect gene expression.
- Polygenic Inheritance: Traits controlled by multiple genes.
- Genetic Drift and Gene Flow: How populations evolve over time.
2. Genetic Disorders
- Down Syndrome: Causes, symptoms, and treatments.
- Cystic Fibrosis: Genetic basis and therapeutic strategies.
- Sickle Cell Anemia: How a single gene mutation causes this disorder.
- Huntington’s Disease: Understanding this neurodegenerative condition.
- Muscular Dystrophy: Genetic causes and potential therapies.
- Hemophilia: The genetics behind this blood disorder.
- Albinism: Genetic mutations leading to a lack of pigmentation.
- Tay-Sachs Disease: Genetic basis and prevention.
- Phenylketonuria (PKU): How diet can manage this genetic disorder.
- BRCA Gene Mutations: Risk factors for breast and ovarian cancer.
3. Molecular Genetics
- DNA Replication: How cells copy their genetic material.
- Gene Cloning: Techniques and applications.
- CRISPR-Cas9: Revolutionizing genetic editing.
- RNA Interference (RNAi): Gene silencing and its applications.
- Transcription and Translation: From DNA to proteins.
- Genetic Recombination: How genes shuffle during reproduction.
- Telomeres and Aging: The role of telomeres in cellular aging.
- MicroRNAs: Small regulators with a big impact.
- Mitochondrial DNA: Inheritance and disorders.
- Exons and Introns: Understanding gene structure.
4. Population Genetics
- Hardy-Weinberg Principle: Predicting allele frequencies.
- Genetic Bottlenecks: How populations lose genetic diversity.
- Founder Effect: When a small population colonizes a new area.
- Genetic Drift: Random changes in allele frequencies.
- Gene Flow: How migration affects genetic variation.
- Natural Selection: Survival of the fittest in genetic terms.
- Genetic Variation in Human Populations: Understanding human diversity.
- Population Stratification: The impact of subgroups on genetic studies.
- Consanguinity: Genetic effects of inbreeding.
- Human Migration Patterns: Tracing ancestry through genetics.
5. Genetics in Medicine
- Pharmacogenomics: Personalized medicine based on genetics.
- Cancer Genetics: How genetic mutations lead to cancer.
- Genetic Counseling: Helping families understand genetic risks.
- Prenatal Genetic Testing: Techniques and ethical considerations.
- Gene Therapy: Correcting genetic defects.
- Stem Cell Research: Potential for treating genetic disorders.
- Precision Medicine: Tailoring treatments to genetic profiles.
- Genetic Markers for Disease: Identifying risk factors.
- Rare Genetic Diseases: Challenges in diagnosis and treatment.
- Genomics in Public Health: Using genetics to track and control diseases.
6. Genetic Engineering and Biotechnology
- Genetically Modified Organisms (GMOs): Benefits and controversies.
- CRISPR Technology: The future of genetic engineering.
- Synthetic Biology: Designing new biological systems.
- Gene Drives: Controlling populations with genetics.
- Cloning: From Dolly the sheep to human cloning debates.
- Genetic Engineering in Agriculture: Improving crop yields and resistance.
- Biotechnology in Medicine: Creating biopharmaceuticals.
- DNA Fingerprinting: Applications in forensics.
- Bioinformatics: Using data to understand genetics.
- Metagenomics: Studying genetic material from environmental samples.
7. Ethics in Genetics
- Designer Babies: The ethics of genetic selection.
- Genetic Privacy: Protecting personal genetic information.
- Eugenics: Historical perspectives and modern implications.
- Ethical Issues in Genetic Testing: Consent, access, and implications.
- Gene Editing Ethics: Where do we draw the line?
- Animal Genetic Modification: Ethical considerations.
- Intellectual Property in Genetics: Patenting genes and their implications.
- Genetic Discrimination: Risks in insurance and employment.
- The Role of Ethics Committees in Genetic Research: Ensuring ethical practices.
- Public Perception of Genetic Technologies: Shaping policy and practice.
8. Evolutionary Genetics
- Molecular Evolution: How DNA changes over time.
- Speciation: The role of genetics in the formation of new species.
- Genetic Evidence for Human Evolution: Tracing our ancestry.
- Adaptive Evolution: How species adapt to their environments.
- Molecular Clocks: Dating evolutionary events.
- Convergent Evolution: Different paths, same traits.
- Genetic Basis of Sexual Selection: How genes influence mating behavior.
- Pleiotropy and Evolution: One gene, multiple effects.
- Co-evolution: How species evolve together.
- Horizontal Gene Transfer: Sharing genes across species.

9. Genetics and Society
- Genetic Ancestry Testing: Personal and societal impacts.
- Genetics and Education: Tailoring learning based on genetics.
- The Genetics of Intelligence: Controversies and findings.
- Genetic Influences on Behavior: Nature vs. nurture debate.
- Forensic Genetics: Solving crimes with DNA.
- Genetic Testing in Sports: Fair play or advantage?
- Gene Patents: Who owns your genes?
- Genetics in the Courtroom: DNA evidence and legal implications.
- Public Understanding of Genetics: Bridging the knowledge gap.
- Genetics in Pop Culture: How genetics is portrayed in media.
10. Emerging Trends in Genetics
- Epigenetic Therapy: Reversing diseases by altering gene expression.
- Single-Cell Genomics: Understanding individual cell functions.
- Gene Editing in Embryos: The future of reproductive technology.
- Personalized Nutrition: Tailoring diets to genetic profiles.
- Biobanks and Big Data: Using large datasets to study genetics.
- Artificial Intelligence in Genomics: Enhancing research with AI.
- Gene Editing for Climate Change: Engineering resilient crops.
- Mitochondrial Replacement Therapy: Preventing genetic diseases.
- CRISPR in Space: Genetic research beyond Earth.
- The Future of Genetic Counseling: Evolving with new technologies.
Tips for Selecting the Right Topic
- Know Your Audience: Tailor your topic complexity based on your audience’s familiarity with genetics. For beginners, stick to fundamental concepts. For more advanced audiences, explore cutting-edge research or ethical debates.
- Purpose of the Presentation: If your goal is to educate, choose topics that explain basic concepts. If you're presenting at a conference, focus on new research or controversial topics.
- Personal Interest: Pick a topic you are passionate about. Your enthusiasm will reflect in your presentation and engage your audience.
- Relevance: Choose topics that are currently in the news or part of ongoing debates in the scientific community.
- Visual Aids: Genetics topics often benefit from visual representations like charts, diagrams, and videos. Choose a topic that lends itself well to these tools.
- Complexity: Ensure that the topic is neither too broad nor too narrow. A broad topic might overwhelm the audience, while a narrow one might not provide enough content for a meaningful presentation. Aim for a balance that allows you to cover the topic comprehensively but concisely.
Final Thoughts
Create ppt using ai.
Just Enter Topic, Youtube URL, PDF, or Text to get a beautiful PPT in seconds. Use the bulb for AI suggestions.
character count: 0 / 6000 (we can fetch data from google)
upload pdf, docx, .png
less than 2 min
Ayan Ahmad Fareedi
writer at MagicSlides
Funny Presentation Topics for Friends: 100+ Hilarious Ideas to Get Everyone Laughing
28 September 2024
100+ Pecha Kucha Presentation Topics to Inspire Your Next Talk
The Ultimate List of Technology Topics for Presentations: 100+ Ideas to Inspire and Engage Your Audience
Easy Topics for Presentation: 100+ Ideas for Students and Professionals
AI Topics for Presentation: 100+ Topics with Categories and Tips for Selection
Military Topics for Presentation: 100+ Ideas with Detailed Explanations
Sociology Topics for Presentation: 100+ Thought-Provoking Ideas
Kid-Friendly Topics for Presentations: 100+ Fun and Engaging Ideas
Stunning presentations in seconds with AI
Install MagicSlides app now and start creating beautiful presentations. It's free!
Get AI-Generated Presentations Ready in Seconds
Free AI PPT Tools
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
The PMC website is updating on October 15, 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Forensic Sci Int Synerg
Recent advances in forensic biology and forensic DNA typing: INTERPOL review 2019–2022
Associated data.
This review paper covers the forensic-relevant literature in biological sciences from 2019 to 2022 as a part of the 20th INTERPOL International Forensic Science Managers Symposium. Topics reviewed include rapid DNA testing, using law enforcement DNA databases plus investigative genetic genealogy DNA databases along with privacy/ethical issues, forensic biology and body fluid identification, DNA extraction and typing methods, mixture interpretation involving probabilistic genotyping software (PGS), DNA transfer and activity-level evaluations, next-generation sequencing (NGS), DNA phenotyping, lineage markers (Y-chromosome, mitochondrial DNA, X-chromosome), new markers and approaches (microhaplotypes, proteomics, and microbial DNA), kinship analysis and human identification with disaster victim identification (DVI), and non-human DNA testing including wildlife forensics. Available books and review articles are summarized as well as 70 guidance documents to assist in quality control that were published in the past three years by various groups within the United States and around the world.
1. Introduction
This review explores developments in forensic biology and forensic DNA analysis of biological evidence during the years 2019–2022. In some cases, there may be overlap with 2019 articles mentioned in the previous INTERPOL review covering 2016 to 2019 [ 1 ]. This review includes books and review articles, published guidance documents to assist in quality control, rapid DNA testing, using law enforcement DNA databases plus investigative genetic genealogy DNA databases along with privacy/ethical issues, forensic biology and body fluid identification, DNA extraction and typing methods, mixture interpretation involving probabilistic genotyping software (PGS), DNA transfer and activity level evaluations, next-generation sequencing (NGS), DNA phenotyping, lineage markers (Y-chromosome, mitochondrial DNA, X-chromosome), new markers and approaches (microhaplotypes, proteomics, and microbial DNA), kinship analysis and human identification with disaster victim identification (DVI), and non-human DNA testing including wildlife forensics.
Multiple searches, using the Scopus (Elsevier) and Web of Science (Clarivate) databases, were conducted in the first half of 2022 with “forensic” and “DNA” or “biology” and “2019 to 2022” as search options. Over 4000 articles were returned with these searches. Through visual examination of titles and authors, duplicates were removed, and articles sorted into 32 subcategories to arrive at a list of almost 2000 publications that were supplemented throughout the remainder of the year as this review was being prepared. The tables of contents for non-indexed journals, such as WIRES Forensic Science , Journal of Forensic Identification , and Forensic Genomics were also examined to locate potentially relevant articles.
For example, a Scopus search conducted on June 13, 2022, using “forensic DNA” and “2019 to 2022” found a total of 3059 documents. Table 1 lists the top ten journals from this search. The Forensic Science International: Genetics Supplement Series (see row #4 in Table 1 ) provides the proceedings of the International Society for Forensic Genetics (ISFG) meeting held in Prague in September 2019. This volume contains 914 pages with 347 articles (although only 172 showed up in the Scopus search) that are freely available at https://www.fsigeneticssup.com /[ 2 ]. Thus, searches conducted with one or even multiple databases (e.g., Scopus and Web of Science) may not be comprehensive or exhaustive.
Top ten journals with forensic DNA articles published from 2019 to 2022 based on a Scopus search on June 13, 2022.
| Ranking | Journal Titles | Number of Articles on Forensic DNA (2019–2022) |
|---|---|---|
| 1 | 429 | |
| 2 | 277 | |
| 3 | 188 | |
| 4 | 172 | |
| 5 | 109 | |
| 6 | 79 | |
| 7 | 65 | |
| 8 | 64 | |
| 9 | 55 | |
| 10 | 52 |
1.1. Books, special issues, and review articles of note
Books published during the period of this review relating to forensic biology and forensic DNA include Essential Forensic Biology, Third Edition [ 3 ], Principles and Practices of DNA Analysis: A Laboratory Manual for Forensic DNA Typing [ 4 ], Forensic DNA Profiling: A Practical Guide to Assigning Likelihood Ratios [ 5 ], Forensic Practitioner's Guide to the Interpretation of Complex DNA Profiles [ 6 ], Silent Witness: Forensic DNA Evidence in Criminal Investigations and Humanitarian Disasters [ 7 ], Mass Identifications: Statistical Methods in Forensic Genetics [ 8 ], Probability and Forensic Evidence: Theory, Philosophy, and Applications [ 9 ], Interpreting Complex Forensic DNA Evidence [ 10 ], Understanding DNA Ancestry [ 11 ], Understanding Forensic DNA [ 12 ], and Handbook of DNA Profiling [ 13 ]. The 2022 Handbook of DNA Profiling spans two volumes and 1206 pages with 54 chapters from 115 contributors representing 17 countries.
Over the past three years, several special issues on topics related to forensic biology were published in Forensic Science International: Genetics and Genes . These special issues were typically collated virtually rather than physically as invited articles were published online over some period of time and then bundled together virtually as a special issue. Some of these review articles or a set of special issue articles are open access (i.e., the authors paid a publication fee so that the article would be available online for free to readers).
During the time frame of this INTERPOL DNA review, FSI Genetics published two special issues: (1) “Trends and Perspectives in Forensic Genetics” (editor: Manfred Kayser) 1 with nine review and two original research articles published between September 2018 and January 2019, and (2) “Forensic Genetics – Unde venisti et quo vadis?” [Latin for “where did you come from and where are you going?”] (editor: Manfred Kayser) with nine articles published in 2021 and early 2022 and likely two more before the end of 2022. Topics for review articles in these special issues include DNA transfer [ 14 ], probabilistic genotyping software [ 15 ], microhaplotypes in forensic genetics [ 16 ], investigative genetic genealogy [ 17 ], forensic proteomics [ 18 ], distinguishing male monozygotic twins [ 19 ], and using the human microbiome for estimating post-mortem intervals and identifying individuals, tissues, or body fluids [ 20 , 21 ]. All of these topics will be discussed later in this article.
A Genes special issue “Forensic Genetics and Genomics” (editors: Emiliano Giardina and Michele Ragazzo) 2 published 11 online articles plus an editorial from April 2020 to January 2021 while another Genes special issue “Forensic Mitochondrial Genomics” (editors: Mitch Holland and Charla Marshall) 3 compiled 11 articles from February 2020 to April 2021. An “Advances in Forensic Genetics” Genes special issue (editor: Niels Morling) 4 included 25 articles shared between April 2021 and May 2022. In July 2022, the Advances in Forensic Genetics articles were compiled as a 518-page book. 5 Other Genes special issues in development or forthcoming covering aspects of forensic DNA and requesting potential manuscripts by late 2022 or early 2023 include “State-of-the-Art in Forensic Genetics” (editor: Chiara Turchi), 6 “Trends in Population Genetics and Identification—Impact on Anthropology (editors: Antonio Amorim, Veronica Gomes, Luisa Azevedo), 7 “Identification of Human Remains for Forensic and Humanitarian Purposes: From Molecular to Physical Methods” (editors: Elena Pilli, Cristina Cattaneo), 8 “Improved Methods in Forensic and DNA Analysis” (editor: Marie Allen), 9 “Forensic DNA Mixture Interpretation and Probabilistic Genotyping” (editor: Michael Coble) 10 , and “Advances in Forensic Molecular Genetics” (editors: Erin Hanson and Claire Glynn). 11 There has been a proliferation of review articles and special issues in this field in the past several years!
A new journal Forensic Science International: Reports was launched in November 2019. As of June 2022, it has published 89 articles involving DNA, most of which are descriptions of population genetic data. Likewise, a June 27, 2022, PubMed search with “forensic DNA” and the journal “Genes” found 88 articles – many of which are part of the previously mentioned special issues.
1.2. Guidance documents
Numerous documentary standards and guidance documents related to forensic DNA have been published by various organizations around the world. Table 2 lists 70 such documents released in the past three years (2019–2022) in the United States, UK, Australia, and the European Union.
Guidance documents related to forensic DNA published from 2019 to 2022. The titles are hyperlinked to available documents. Abbreviations: FBI (Federal Bureau of Investigation), CODIS (Combined DNA Index System), SWGDAM (Scientific Working Group on DNA Analysis Methods), NGS (next generation sequencing), US DOJ (United States Department of Justice), ULTR (Uniform Language for Testimony and Reports), AABB (Association for the Advancement of Blood and Biotherapies), ASB (Academy Standards Board), OSAC (Organization of Scientific Area Committees for Forensic Science), UKFSR (United Kingdom Forensic Science Regulator), ENFSI (European Network of Forensic Science Institutes), NIFS (National Institute of Forensic Science), ISFG (International Society for Forensic Genetics).
| Organization | Publication Date | Guidance Document Title |
|---|---|---|
| FBI | July 2020 | Quality Assurance Standards for Forensic DNA Testing Laboratories |
| FBI | July 2020 | Quality Assurance Standards for DNA Databasing Laboratories |
| FBI | July 2020 | Quality Assurance Standards Audit for Forensic DNA Testing Laboratories |
| FBI | July 2020 | Quality Assurance Standards Audit for DNA Databasing Laboratories |
| FBI | July 2020 | Guidance Document for the FBI Quality Assurance Standards for Forensic DNA Testing and DNA Databasing |
| FBI | Jan 2022 | A Guide to All Things Rapid DNA (13 pages; see also Hares et al., 2020 [ ]) |
| FBI | Sept 2019 | Non-CODIS Rapid DNA Considerations and Best Practices for Law Enforcement Use (7 pages) |
| FBI | July 2020 | Rapid DNA Testing for non-CODIS uses: Considerations for Court (5 pages) |
| SWGDAM | Apr 2019 | Mitochondrial DNA Analysis Revisions Related to NGS |
| SWGDAM | Apr 2019 | Addendum to Interpretation Guidelines to Address NGS |
| SWGDAM | Feb 2020 | Overview of Investigative Genetic Genealogy |
| SWGDAM | July 2020 | Report on Y-Screening of Sexual Assault Evidence Kits (SAEKs) |
| SWGDAM | July 2020 | Training Guidelines |
| SWGDAM | Jan 2022 | YHRD Updates for U.S. Laboratories |
| SWGDAM | Mar 2022 | Interpretation Guidelines for Y-Chromosome STR Typing by Forensic DNA Laboratories |
| SWGDAM | Mar 2022 | Supplemental Information for the SWGDAM Interpretation Guidelines for Y-Chromosome STR Typing by Forensic DNA Laboratories |
| US DOJ | July 2019 | Bureau of Justice Assistance (BJA) Triage of Forensic Evidence Testing: A Guide for Prosecutors (49 pages) |
| US DOJ | May 2022 | National Institute of Justice (NIJ) National Best Practices for Improving DNA Laboratory Process Efficiency (104 pages) |
| US DOJ | Mar 2019 | Approved ULTR for the Forensic DNA Discipline – Autosomal DNA with Probabilistic Genotyping (5 pages) |
| US DOJ | Mar 2019 | Approved ULTR for the Forensic DNA Discipline – Mitochondrial DNA (4 pages) |
| US DOJ | Mar 2019 | Approved ULTR for the Forensic DNA Discipline – Y-STR DNA (4 pages) |
| US DOJ | Nov 2019 | Interim Policy on Forensic Genetic Genealogical DNA Analysis and Searching (8 pages [ ]; see also Callaghan 2019 [ ]) |
| US DOJ | Dec 2019 | Needs Assessment of Forensic Laboratories and Medical Examiner/Coroner Offices: Report to Congress (200 pages) |
| US DOJ | Sept 2021 | NIJ Forensic Laboratory Needs Technology Working Group (FLN-TWG) Implementation Strategies: Next Generation Sequencing for DNA Analysis (29 pages) |
| US DOJ | May 2022 | A Landscape Study Examining Technologies and Automation for Differential Extraction and Sperm Separation for Sexual Assault Investigations (50 pages) |
| US DOJ | Sept 2022 | An Introduction to Forensic Genetic Genealogy Technology for Forensic Science Service Providers (7 pages) |
| ASB | Aug 2019 | Standard for Forensic DNA Analysis Training Programs (ANSI/ASB 022) |
| ASB | Sept 2019 | Standard for Forensic DNA Interpretation and Comparison Protocols (ANSI/ASB 040) |
| ASB | June 2020 | Standard for Training in Forensic DNA Isolation and Purification Methods (ANSI/ASB 023) |
| ASB | July 2020 | Standard for Validation of Probabilistic Genotyping Systems (ANSI/ASB 018) |
| ASB | Aug 2020 | Standard for Internal Validation of Forensic DNA Analysis Methods (ANSI/ASB 038) |
| ASB | Aug 2020 | Standards for Training in Forensic Serological Methods (ANSI/ASB 110) |
| ASB | Aug 2020 | Standard for Training in Forensic Short Tandem Repeat Typing Methods using Amplification, DNA Separation, and Allele Detection (ANSI/ASB 115) |
| ASB | Aug 2020 | Standard for Training in Forensic DNA Quantification Methods (ANSI/ASB 116) |
| ASB | Sept 2020 | Standard for the Developmental and Internal Validation of Forensic Serological Methods (ANSI/ASB 077) |
| ASB | May 2021 | Standard for Training in Forensic DNA Amplification Methods for Subsequent Capillary Electrophoresis Sequencing (ANSI/ASB 130) |
| ASB | Aug 2021 | Standard for Training in Forensic DNA Sequencing using Capillary Electrophoresis (ANSI/ASB 131) |
| ASB | Sept 2021 | Standard for Training in Forensic Human Mitochondrial DNA Analysis, Interpretation, Comparison, Statistical Evaluation, and Reporting (ANSI/ASB 140) |
| OSAC | Mar 2020 | Human Factors in Validation and Performance Testing of Forensic Science (35 pages) |
| OSAC | Apr 2021 | Best Practice Recommendations for the Management and Use of Quality Assurance DNA Elimination Databases in Forensic DNA Analysis (OSAC 2020-N-0007) |
| OSAC | June 2021 | Standard for Interpreting, Comparing and Reporting DNA Test Results Associated with Failed Controls and Contamination Events (OSAC 2020-S-0004) |
| OSAC | May 2022 | Human Forensic DNA Analysis (Current Practice) Process Map (42 pages) |
| UKFSR | Mar 2021 | FSR-C-100, Issue 7 – Codes of Practice and Conduct (2021) |
| UKFSR | Sept 2020 | FSR-C-108, Issue 2 – DNA Analysis: Codes of Practice and Conduct |
| UKFSR | May 2020 | FSR-C-116, Issue 1 – Sexual Assault Examination: Requirements for the Assessment, Collection and Recording of Forensic Science Related Evidence |
| UKFSR | Jan 2021 | FSR-C-118, Issue 1 – Development of Evaluative Opinions |
| UKFSR | Sept 2020 | FSR-G-201, Issue 2 – Validation |
| UKFSR | Sept 2020 | FSR-G-202, Issue 2 – The Interpretation of DNA Evidence (Including Low-Template DNA) |
| UKFSR | Sept 2020 | FSR-P-300, Issue 2 – Validation – Use of Casework Material |
| UKFSR | Sept 2020 | FSR-P-302, Issue 2 – DNA Contamination Detection: The Management and Use of Staff Elimination DNA Databases |
| UKFSR | Sept 2020 | FSR-G-206, Issue 2 – The Control and Avoidance of Contamination in Scene Examination involving DNA Evidence Recovery |
| UKFSR | Sept 2020 | FSR-G-207, Issue 2 – The Control and Avoidance of Contamination in Forensic Medical Examinations |
| UKFSR | Sept 2020 | FSR-G-208, Issue 2 – The Control and Avoidance of Contamination in Laboratory Activities involving DNA Evidence Recovery Analysis |
| UKFSR | May 2020 | FSR-G-212, Issue 1 – Guidance for the Assessment, Collection and Recording of Forensic Science Related Evidence in Sexual Assault Examinations |
| UKFSR | Sept 2020 | FSR-G-213, Issue 2 – Allele Frequency Databases and Reporting Guidance for the DNA (Short Tandem Repeat) Profiling |
| UKFSR | Sept 2020 | FSR-G-217, Issue 2 – Cognitive Bias Effects Relevant to Forensic Science Examinations |
| UKFSR | Sept 2020 | FSR-G-222, Issue 3 – DNA Mixture Interpretation |
| UKFSR | Sept 2020 | FSR-G-223, Issue 2 – Software Validation for DNA Mixture Interpretation |
| UKFSR | Jun 2020 | FSR-G-224, Issue 1 – Proficiency Testing Guidance for DNA Mixture Analysis and Interpretation |
| UKFSR | Mar 2021 | FSR-G-227, Issue 1 – Y-STR Profiling |
| UKFSR | Apr 2021 | FSR-G-228, Issue 1 – DNA Relationship Testing using Autosomal Short Tandem Repeats |
| UKFSR | Apr 2021 | FSR-G-229, Issue 1 – Methods Employing Rapid DNA Devices |
| ENFSI DNA | Apr 2019 | DNA Database Management Review and Recommendations |
| ENFSI DNA | Mar 2022 | Guideline for the Training of Staff in Forensic DNA Laboratories |
| NIFS | Sept 2019 | Case Record Review in Forensic Biology |
| NIFS | Sept 2019 | Empirical Study Design in Forensic Science - A Guideline to Forensic Fundamentals |
| NIFS | Dec 2019 | Transitioning Technology from the Laboratory to the Field - Process and Considerations for the Forensic Sciences |
| AABB | Jan 2022 | Standards for Relationship Testing Laboratories, 15th Edition |
| ISFG DNA Commission | Jan 2020 | Assessing the value of forensic biological evidence – Guidelines highlighting the importance of propositions. Part II: Evaluation of biological traces considering activity level propositions (Gill et al., 2020 [ ]) |
| ISFG DNA Commission | June 2020 | Recommendations on the interpretation of Y-STR results in forensic analysis (Roewer et al., 2020 [ ]) |
1.2.1. SWGDAM, FBI, and other US DOJ activities
The Federal Bureau of Investigation (FBI) Laboratory funds the Scientific Working Group on DNA Analysis Methods (SWGDAM) 12 to serve as a forum for discussing, sharing, and evaluating forensic biology methods, protocols, training, and research. In addition to creating guidelines on various topics, SWGDAM, which meets semiannually in January and July, provides recommendations to the FBI Director on the Quality Assurance Standards (QAS) used to assess U.S. forensic DNA laboratories involved in the National DNA Index System (NDIS) that perform DNA databasing and forensic casework. New versions of the QAS became effective July 1, 2020.
SWGDAM work products from the timeframe of 2019–2022 (see Table 2 ) include QAS audit and guidance documents, mitochondrial DNA analysis and short tandem repeat (STR) interpretation guideline revisions related to next-generation sequencing (NGS), training and Y-chromosome interpretation guidelines, a Y-chromosome Haplotype Reference Database (YHRD) update for U.S. laboratories, and reports on investigative genetic genealogy and Y-screening of sexual assault evidence kits. These documents are all accessible online. 13
In January 2022, the FBI produced a 13-page guide 14 on rapid DNA testing describing booking station applications and their vision for future integration of crime scene sample analysis and the Combined DNA Index System (CODIS), which builds on a joint position statement published in July 2020 by leaders of U.S. and European groups [ 22 ]. In addition, the FBI has shared guidance on their website for non-CODIS use of rapid DNA testing with law enforcement applications 15 and considerations for court. 16
United States Department of Justice (US DOJ) Uniform Language for Testimony and Reports (ULTRs), 17 contain three ULTRs for the forensic DNA discipline that became effective in March 2019: autosomal DNA with probabilistic genotyping, mitochondrial DNA, and Y-STR DNA. USDOJ also released an interim policy on investigative genetic genealogy in November 2019 [ 23 ] along with an opinion piece in the journal Science calling for responsible genetic genealogy [ 24 ].
Other agencies within US DOJ, namely the Bureau of Justice Assistance (BJA) and the National Institute of Justice (NIJ), published a guide for prosecutors on triaging forensic evidence [ 25 ] and best practices for improving DNA laboratory process efficiency [ 26 ]. A 200-page report to Congress on the needs assessment of forensic laboratories and medical examiner/coroner offices was released in December 2019 calling for $640 million annually in additional funding to support U.S. forensic efforts [ 27 ].
In September 2021, the Forensic Technology Center of Excellence (FTCOE), which is funded by NIJ, published a 29-page implementation strategy on next-generation sequencing for DNA analysis that was written by the NIJ Forensic Laboratory Needs Technology Working Group (FLN-TWG) [ 28 ]. In May 2022, FTCOE released a 50-page landscape study examining technologies and automation for differential extraction and sperm separation used in sexual assault investigations [ 29 ]. An introduction to forensic genetic genealogy was released in September 2022 [ 30 ].
The FTCOE also published a human factors forensic science sourcebook 18 in March 2022 through open access articles in the journal Forensic Science International: Synergy . This sourcebook, which has general applicability rather than being specific to forensic DNA analysts, includes an overview article [ 31 ] along with articles on personnel selection and assessment [ 32 ], the benefits of committing errors during training [ 33 ], how characteristics of human reasoning and certain situations can contribute to errors [ 34 ], stressors that impact performance [ 35 ], and the impact of communication between forensic analysts and detectives using a new metaphor [ 36 ].
1.2.2. OSAC and ASB activities
The Organization of Scientific Area Committees for Forensic Science (OSAC) 19 is congressionally-funded and administered by the Special Programs Office within the National Institute of Standards and Technology (NIST). OSAC consists of a governing board and over 600 members and associates organized into seven scientific area committees (SACs) and 22 subcommittees. The Biology SAC is divided into human and wildlife forensic biology activities. The Human Forensic Biology Subcommittee 20 focuses on standards and guidelines related to training, method development and validation, data analysis, interpretation, and statistical analysis as well as reporting and testimony for human forensic serological and DNA testing. The Wildlife Forensics Subcommittee 21 works on standards and guidelines related to taxonomic identification, individualization, and geographic origin of non-human biological evidence based on morphological and genetic analyses.
The Academy Standards Board (ASB) 22 is a wholly owned subsidiary of the American Academy of Forensic Sciences (AAFS) and was established as a standards developing organization (SDO). In 2015, ASB was accredited as an SDO by the American National Standards Institute (ANSI). The ASB DNA Consensus Body, with a membership consisting of practitioners, researchers, and lawyers, develops standards and guidelines related to the use of DNA in legal proceedings. Many of the documents developed by ASB were originally proposed OSAC standards or guidelines.
The OSAC Registry 23 is a repository of high-quality and technically-sound standards (both published and proposed) that are intended for implementation in forensic science laboratories. As of July 2022, the OSAC Registry contains 11 standards published by ASB as well as two (2) proposed OSAC standards or best practice recommendations related to human forensic biology. Another four ASB standards and two proposed OSAC standards related to wildlife forensic biology are on the OSAC Registry. The ASB standards issued in the past three years related to human forensic biology cover interpretation and comparison protocols, training in various parts of the process, and validation of forensic serological and DNA analysis methods as well as probabilistic genotyping systems (see Table 2 for names of these documents). A number of other documents 24 related to serological testing methods, assigning propositions for likelihood ratios in forensic DNA interpretations, validation of forensic DNA methods and software, familial DNA searching, management and use of quality assurance DNA elimination databases, setting thresholds, evaluative forensic DNA testimony, and training in use of statistics are in development within OSAC and ASB.
Additional work products of OSAC include (1) a lexicon 25 with 3282 records (although multiple records may exist for the same word, e.g., there are five definitions provided for “validation” from various sources), (2) a 35-page technical guidance document 26 on human factors in validation and performance testing that describes key issues in designing, conducting, and reporting validation research, (3) a listing of research and development needs in forensic science 27 including 18 identified by the OSAC Human Forensic Biology Subcommittee during their deliberations ( Table 3 ), and (4) process maps for several forensic disciplines including a 42-page depiction of current practices and decisions in human forensic DNA analysis released in May 2022 [ 37 ]. As a visual representation of critical steps and decision points, a process map is intended to help improve efficiencies and reduce errors, and highlight gaps where further research or standardization would be beneficial. Process maps can assist with training new examiners and enable development of specific laboratory policies or help identify best practices for the field.
Research and development needs in forensic biology as identified by the OSAC Human Forensic Biology Subcommittee (as of July 2022, see https://www.nist.gov/osac/osac-research-and-development-needs ).
| OSAC Listed R&D Needs | |
|---|---|
| 1 | Applications of the Microbiome in DNA Transfer and Human Identification |
| 2 | Assessing DNA Background and Transfer Scenarios in Forensic Casework |
| 3 | Best Practices to Minimize Potential Biases in the Generation and Interpretation of DNA Profiles |
| 4 | Best Practices for Reporting Likelihood Ratios or Other Probabilistic Results in Court |
| 5 | Characterization, Development and Validation of Methods in Single Cell Isolation and Analysis |
| 6 | Characterization, Optimization and Comparison of DNA Sequencing Methods |
| 7 | Characterizing the Presence and Prevalence of Cell-Free DNA |
| 8 | Development of Infrastructure to Compile and Share Raw Electronic Data for Training and Tool Development |
| 9 | Efficiency, Throughput and Speed Improvements in Rapid DNA Instrumentation Through the Development of Direct PCR Methods |
| 10 | Efficient Collection of DNA at the Scene and from Evidence Items |
| 11 | Establishing the Value and Designing a Process for Including Flanking Region SNPs in Massive Parallel Sequencing Based on STRP Casework |
| 12 | Improving the Recovery of Male DNA from Sexual Assault Kits |
| 13 | Methods in Forensic Genealogy |
| 14 | Non-PCR Based Methods for DNA Amplification and/or Detection |
| 15 | Optimization of DNA Extraction for Low Level Samples |
| 16 | Software Solutions for Low Template and High Order DNA Mixture Interpretation in Sequence and Fragment-Based Methods |
| 17 | Software Solutions for Y-STR Mixture Deconvolution |
| 18 | Solutions in Phenotyping and Ancestry Analyses |
1.2.3. UK Forensic Science Regulator
The UK Forensic Science Regulator (UKFSR) oversees forensic science efforts in England, Wales, and Northern Ireland. In March 2021, the Regulator released the seventh issue 28 of the Codes of Practice and Conduct for forensic science providers and practitioners in the criminal justice system. This 114-page document, which has been updated every few years, provides the overall framework for forensic science activities in the UK with other supporting guidance documents on specific areas like DNA analysis or general tasks like validation. In September 2020, a number of the Regulator documents were revised and reissued. As noted in Table 2 (see rows with documents containing “Issue 1” in the title), new guidance documents were also released in the past few years on sexual assault examinations, development of evaluative opinions, proficiency testing for DNA mixture interpretation, Y-STR profiling, DNA relationship testing, and methods employing rapid DNA testing devices. Table 2 lists 20 guidance documents pertinent to forensic biology from the UKFSR.
1.2.4. European Union and Australia
The European Network of Forensic Science Institutes (ENFSI) DNA Working Group published two documents in the past three years: one on DNA database management and the other on training of staff in forensic DNA laboratories (see Table 2 ). A best practice manual for human forensic biology and DNA profiling is also under development.
The Australian National Institute of Forensic Science (NIFS) published three documents of relevance to forensic biology on case record review, empirical study design, and transitioning technology from the laboratory to the field (see Table 2 ).
1.2.5. Other international efforts
The Association for the Advancement of Blood and Biotherapies (AABB) 29 published the 15th edition of their Standard for Relationship Testing Laboratories, which became effective on January 1, 2022. This documentary standard was developed by the AABB Relationship Testing Standards Committee and applies to laboratories accredited for paternity testing and other forms of genetic relationship assessment.
The International Society for Forensic Genetics (ISFG) DNA Commission 30 published two articles during the timeframe of this INTERPOL review (see Table 2 ). In 2020, guidelines and considerations were published on evaluating DNA results under activity level propositions [ 38 ]. In addition, the state of the field regarding interpretation of Y-STR results was examined along with different approaches for haplotype frequency estimation using population data – with the Discrete Laplace approach being recommended [ 39 ]. Future ISFG DNA Commission efforts will address STR allele sequence nomenclature and phenotyping.
2. Advancements in current practices
This section (Section 2 ) is intended to be law enforcement and practitioner-focused through examination of advances in current practices. The following section (Section 3 ) is intended to be researcher-focused through emphasis on emerging technologies and new developments. In this section, topics specifically covered include rapid DNA analysis, use of DNA databases to aid investigations (including familial searching, investigative genetic genealogy, genetic privacy and ethical concerns, and sexual assault kit testing), body fluid identification, DNA extraction and typing methods, and DNA interpretation at the sub-source and activity level.
2.1. Rapid DNA analysis
Rapid DNA instruments that provide integrated “swab-in-profile-out” results in 90 min or less can be used in police booking station environments and assist investigations outside of a traditional laboratory environment. These instruments were initially designed for analysis of buccal swabs to help speed processing of reference samples associated with criminal cases. Such samples are expected to contain relatively large quantities of DNA from a single contributor. Some attempts to extend the range of sample types to low quantities of DNA or mixtures have been published with various levels of success (see Table 4 ). Researcher and practitioners from Australia [ [40] , [41] , [42] ], Canada [ 43 ], China [ 44 ], Italy [ 45 ], Japan [ 46 , 47 ], and the United States [ [48] , [49] , [50] , [51] , [52] , [53] , [54] , [55] , [56] , [57] ] have contributed to an increased understanding of rapid DNA testing capabilities and limitations.
Summary of 20 rapid DNA instrument validation and evaluation studies published from 2019 to 2022. Abbreviations: A-Chip (arrestee cartridge, designed for high-quantity DNA samples), I-Chip (investigative cartridge, designed for low-quantity DNA samples), ACE (arrestee cartridge with GlobalFiler STR markers), RapidINTEL (uses 32 rather than 28 PCR cycles to increase success with low-quantity DNA samples). A-Chip and I-Chip amplify the FlexPlex set of 23 autosomal STRs, three Y-STRs, and amelogenin [ 51 ]. ACE and RapidINTEL utilize the GlobalFiler set of 21 autosomal STRs, one Y-STR, one Y-chromosome InDel, and amelogenin.
| Publication | Instrument | Cartridge/Kit | Test Performed and Success Rates Reported |
|---|---|---|---|
| Amick & Swiger 2019 [ ] | RapidHIT ID | ACE and EXT | Performed SWGDAM internal validation studies including known and database-type samples, reproducibility, precision, sensitivity, stochastic effects, mixtures, contamination assessment, and concordance studies |
| Carney et al., 2019 [ ] | ANDE 6C | A-Chip | Conducted SWGDAM developmental validation (across 6 labs, 2045 swabs, 13 instruments): species specificity, limit of detection, stability, inhibitors, reproducibility, reference material, mixtures, precision, concordance, signal strength, peak height ratio, stutter, non-template addition, resolution, and contamination assessment; ; successfully interpreted >2000 samples with over 99.99% concordant alleles; data package led to receiving NDIS approval in June 2018 |
| Shackleton et al., 2019 [ ] | RapidHIT ID | NGM SElect Express | Described development studies that included process optimization, sensitivity, repeatability, contamination checks, inhibition, swab age, concordance, and overall performance; gave a full profile |
| Shackleton et al., 2019 [ ] | RapidHIT 200 | NGM SElect Express | Performed some protocol adjustments that enhanced slightly the sensitivity with mock crime scene samples (dilutions of blood and cell line DNA) |
| Yang et al., 2019 [ ] | MiDAS | PowerPlex ESI 16 Plus | Described protocols for analysis of reference samples with a fully automated integrated microfluidic system (MiDAS), which is not commercially available |
| Romsos et al., 2020 [ ] | ANDE 6C, RapidHIT ID, RapidHIT 200 | A-Chip, ACE | Reported results from the July 2018 rapid DNA maturity assessment with multiple instruments organized by NIST; the average success rate for obtaining the 20 CODIS core loci was |
| Manzella & Moreno 2020 [ ] | ANDE 6C | A-Chip | Reported success rates on 54 samples of , , like those that may be received during casework processing; with manual interpretation, the CODIS 20 success rate increased to 63% |
| Murakami et al., 2020 [ ] | RapidHIT ID | ACE and RapidINTEL | Examined blood and nail clippings from postmortem bodies with varying degrees of decomposition and reported “the device is useful for samples of sufficient quantity and purity, considering post-mortem intervals of up to approximately one week” |
| Ragazzo et al., 2020 [ ] | ANDE 6C | A-Chip | Compared results for 104 buccal swabs with rapid and conventional protocols, observed a and , and concluded “the ANDE 6C System is robust, reliable, and is suitable for use in human identification for forensic purposes from a single source of DNA” |
| Kitayama et al., 2020 [ ] | ANDE 6C | A-Chip and I-Chip | Examined 19 mock DVI samples; “success rates of putrefied DVI samples varied widely between 0% and 20% and 50%–80% depending on cartridge and sample types” and “DVI samples that yielded more than 1 ng/μL of DNA when extracted with conventional protocols were suitable” (success defined as at least 20 CODIS STRs for A-Chip and any 12 out of 20 CODIS STRs for I-Chip) |
| Turingan et al., 2020 [ ] | ANDE 6C | I-Chip | Processed 1705 mock crime scene and DVI samples across 17 sample types; with 1299 samples in the accuracy study, (defined by the authors as at least 16 CODIS STRs on the first run) |
| Turingan et al., 2020 [ ] | ANDE 6C | A-Chip and I-Chip | Examined tissues and bones from 10 sets of human remains exposed above ground for up to one year; analysis of bone and teeth works best with extended exposure |
| Chen et al., 2021 [ ] | RapidHIT ID | RapidINTEL | Performed substrate, sensitivity, precision, contamination, mock inhibition, mixture, concordance, species, and versatility studies; 100% concordance with conventional CE-based DNA analysis across 19 STRs; |
| Hinton et al., 2021 [ ] | ANDE 6C | A-Chip and I-Chip | Examined in a technical exploitation workflow; |
| Manzella et al., 2021 [ ] | ANDE 6C | I-Chip | Examined 7 muscle tissue, 4 pulverized bone exemplars, 9 rib, and 26 teeth samples and concluded “the robustness and consistency of the method still have room for improvement” |
| Martin et al., 2022 [ ] | RapidHIT ID | RapidINTEL | Examined 8 touched samples (10 replicates each) containing low quantities of DNA; found that the method “was not suitable for the 12G cartridge, insulated wire, or twine sampling in its current form” |
| Ridgley & Olson 2022 [ ] | ANDE 6C | I-Chip | Evaluated a protocol for sexual assault samples; “met the instrument metrics for success and resulted in at least a partial profile” (>8 loci) and “could be immediately used without further review” |
| Cihlar et al., 2022 [ ] | RapidHIT ID | ACE | Performed validation experiments with concordance, contamination, sensitivity, repeatability, reproducibility, swab reprocessing, stability, inhibition, and mixture studies (253 samples total); |
| Ward et al., 2022 [ ] | RapidHIT ID | RapidINTEL | Assessed performance for mixture interpretation using STRmix v2.8 (can yield orders of magnitude different LR values compared to standard laboratory workflow) |
| Watherston et al., 2022 [ ] | ANDE 6C and RapidHIT ID | I-Chip and RapidINTEL | Used donated cadavers with a simulated building collapse scenario; allele recovery varied by sample type and instrument; concluded “further optimization is recommended for highly decomposed and skeletonized human remains” |
The Accelerated Nuclear DNA Equipment (ANDE) 6C (ANDE, Longmont, CO, USA) and the RapidHIT ID (Thermo Fisher Scientific, Waltham, MA, USA) are the current 31 commercially available rapid DNA systems. Each system consists of a swab for introducing the sample, a cartridge or biochip with pre-packed reagents, the instrument, and analysis software with an expert system for automated STR allele calling. Different sample cartridges can be run on each system depending on the sample type and expected quantity of DNA.
For ANDE, the arrestee cartridge (A-Chip), can accommodate up to five samples and is intended for relatively high quantities of DNA typically collected from reference buccal swabs, while the investigative cartridge (I-Chip), can process up to four samples and is intended for lower quantities of DNA that might be present in casework or disaster victim identification samples. Both ANDE cartridges use the FlexPlex27 STR assay that tests 23 autosomal STR loci, three Y-chromosome STRs, and amelogenin to generate data compatible with DNA databases around the world [ 51 ]. The RapidHIT ID ACE cartridge and RapidINTEL cartridge serve similar purposes as the ANDE A-Chip and I-Chip using GlobalFiler Express kit markers (21 autosomal STRs, DYS391, a Y-chromosome insertion/deletion marker, and amelogenin) instead of the FlexPlex assay. The ACE sample cartridge uses buccal swabs while the EXT sample cartridge processes DNA extracts [ 56 ]. Sensitivity is enhanced in the RapidINTEL cartridge by increasing the number of PCR cycles from 28 to 32 and decreasing the lysis buffer volume from 500 μL to 300 μL compared to the ACE cartridge parameters [ 46 ].
With rapid DNA testing's swab-in and answer-out integrated configuration, limited options exist for testing conditions (e.g., either A-Chip or I-Chip with ANDE). Therefore, users should evaluate performance for the sample types they desired to routinely test in their specific environment. Table 4 summarizes recently published studies containing rapid DNA assessments.
National DNA Index System (NDIS) approval has been provided by the FBI Laboratory for accredited forensic DNA laboratories to use either the ANDE 6C or RapidHIT ID Systems (A-Chip and ACE cartridges only) 32 with eligible reference mouth swabs. As noted in Table 2 , the FBI.gov website contains three documents related to rapid DNA testing: “Non-CODIS Rapid DNA Considerations and Best Practices for Law Enforcement Use” (7-pages), “Rapid DNA Testing for Non-CODIS Uses: Considerations for Court” (5-pages), and “A Guide to All Things Rapid DNA” (13-pages) in January 2022 to provide information on the topic to law enforcement agencies.
The ENFSI DNA Working Group, SWGDAM, and an FBI Rapid DNA Crime Scene Technology Advancement Task Group co-published a position statement on the use of rapid DNA testing from crime scene samples [ 22 ]. These groups emphasized the need to have future rapid DNA systems with (1) methods to identify low quantity, degradation, and inhibition as well as meeting the human quantification requirements shared by SWGDAM and others, (2) the ability to export analyzable raw data for analysis or reanalysis by trained and qualified forensic DNA analysts, (3) an on-board fully automated expert system to accurately flag single-source or mixture DNA profiles requiring analyst evaluation, (4) improved peak height ratio balance (per locus and across loci) for low-quality and mixture samples “through enhancements in extraction efficiencies, changes in cycling parameters, and/or changes in STR kit chemistries,” and (5) published developmental validation studies on a wide variety of forensic evidence type samples with “data-supported recommendations regarding types of forensic evidence that are suitable and unsuitable for use with Rapid DNA technology” [ 22 ].
With a likely increase in the capabilities and the availability of rapid DNA systems, investigators will need to decide whether to use this capability onsite in specific situations or to send collected samples to a conventional forensic laboratory for processing at a later time. A group in the Netherlands collaborated with the New York City Police Department Crime Scene Unit and Evidence Collection Team to explore a decision support system [ 60 ]. In this study, participants were informed that rapid DNA testing was less sensitive compared to laboratory analysis and that the sample would be consumed, but that results from rapid DNA testing could identify a suspect within 2 h as opposed to waiting an average of 45 days for the laboratory results [presumably due to sample backlogs]. They were also told that a DNA profile obtained with rapid DNA would be acceptable in court. In the end, “>90% of the participants (85 out of 91) saw added value for using a Rapid DNA device in their investigative process …” with “a systematic approach, which consists of weighing all possible outcomes before deciding to use a Rapid DNA analysis device” [ 60 ]. The authors note that for such an approach to be successful “knowledge on DNA success rates [with various evidence types] is necessary in making evidence-based decisions for Rapid DNA analysis” [ 60 ].
A group in Australia performed a cost-benefit analysis of a decentralized rapid DNA workflow that might exist in the future with instruments placed at police stations around their country [ 61 ]. A virtual assessment considered all reference DNA samples collected during a two-month time period at 10 participating police stations in five regions of Australia. Processing times at the corresponding DNA analysis laboratories were calculated based on when the sample was received compared to the day when a DNA profile was obtained for that sample. From the survey conducted, it was estimated that up to 80,000 reference DNA samples are currently processed each year in forensic DNA laboratories across Australia [ 61 ].
Consumable costs for conventional DNA testing reagents in Australia were found to range from $17 to $35 whereas the rapid DNA consumable costs were estimated to be $100 per sample along with an anticipated $100,000 instrument cost per police station. Of course, the rate of use is expected to vary based on the number of reference samples collected in that jurisdiction. Since rapid DNA instruments utilize consumable cartridges with expiration dates, it was estimated that a police station would need to process six DNA samples per week to avoid having to discard an expired cartridge and thus increase the overall cost of their rapid DNA testing efforts. The authors of this study conclude “that routine laboratory DNA analysis meets the current needs for the majority of cases … It is anticipated that while the cost discrepancy between laboratory and rapid DNA processing remains high, the uptake of the technology in Australia will be limited [at least for a police booking station scenario]” [ 61 ].
Rapid DNA technology can be used in a variety of contexts including some that extend beyond traditional law enforcement. Seven distinct use contexts for rapid DNA capabilities have been described [ 62 ]: (1) evidence processing at or near crime scenes to generate leads for confirmation by a forensic laboratory, (2) booking or detection stations to compare an individual's DNA profile to a forensic database while the individual is still in custody, (3) disaster victim identification to permit rapid DNA processing of a victim's family members during their visit to family assistance centers when filing missing persons reports, (4) missing persons investigations to quickly process unidentified human remains and/or family reference samples to generate leads for confirmation by a forensic laboratory, (5) border security to develop DNA data from detainees for comparison to indices of prior border crossers while the individual is still in custody, (6) human trafficking and immigration fraud detection to permit immigration officials to verify family relationship claims, and (7) migrant family reunification to allow immigration officials to verify parentage claims and reunite family members separated at the border. Social and ethical considerations have been proposed for each of these use contexts in terms of data collection, data access and storage, and oversight and data protection [ 62 ].
One study [ 47 ] evaluating buccal swabs and mock disaster victim identification samples drew an important conclusion worth repeating here: “The Rapid DNA system provides robust and automated analysis of forensic samples without human review. Sample analysis failure can happen by chance in both the Rapid DNA system and conventional laboratory STR testing. While re-injection of PCR product is easily possible in the conventional method, this is not an option with the Rapid DNA system. Accordingly, the Rapid DNA system is a suitable choice but should be limited to samples that can easily be collected again if necessary or to samples that are of sufficient amount for repeated analysis. Application of this system to valuable samples such as those related to casework need to be considered carefully before analysis.”
2.2. Using DNA databases to aid investigations (national databases, familial searching, investigative genetic genealogy, genetic privacy & ethical concerns, sexual assault kit testing)
Forensic DNA databases can aid investigations by demonstrating connections between crime scenes, linking a previously enrolled DNA profile from an arrestee or convicted offender to biological material recovered from a crime scene, or aiding identification of missing persons through association of remains with biological relatives. Establishment of these databases requires significant investments over time to enroll data from crime scenes and potential serial offenders or unidentified human remains and relatives of missing persons. This section explores issues around national DNA databases, familial searching, investigative genetic genealogy, and genetic privacy and ethical concerns.
A systematic review regarding the effectiveness of forensic DNA databases looked at 19 articles published between 1985 and 2018 and found most studies support the assumption that DNA databases are an effective tool for the police, society, and forensic scientists [ 63 ]. Recommendations have been proposed to make cross-border exchange of DNA data more transparent and accountable with the Prüm system that enables information sharing across the European Union [ 64 ]. An analysis of news articles discussing the use of DNA testing in family reunification with migrants separated at the U.S.-Mexico border has been performed [ 65 ], and a standalone humanitarian DNA identification database has been proposed [ 66 ]. Aspects of international DNA kinship matching were explored to aid missing persons investigations and disaster victim identification processes [ 67 ]. A business case was presented for expanded DNA indirect matching using additional genetic markers, such as Y-chromosome STRs, mitochondrial DNA, and X-chromosome STRs, to reveal previously undetected familial relationships [ 68 ].
Approaches to transnational exchange of DNA data include (1) creation of an international DNA database, (2) linked or networked national DNA databases, (3) request-based exchange of data, and (4) a combination of these [ 69 ]. For example, the INTERPOL DNA database 33 contains more than 247,000 profiles contributed by 84 member countries. The I-Familia global database assists with missing persons identification based on international DNA kinship matching. 34
2.2.1. National DNA databases
Since the United Kingdom launched the first national DNA database in 1995, national DNA databases continue to be added in many countries including Brazil [ 70 , 71 ], India [ 72 ], Pakistan [ 73 , 74 ], Portugal [ 75 ], and Serbia [ 76 ]. A survey of 15 Latin American countries found that 13 of them had some kind of DNA database [ 77 ]. The opinions of 210 prisoners and prison officials in three Spanish penitentiary centers were also collected regarding DNA databases [ 78 ].
The effectiveness of databases has been debated over the years. Seven key indicators were used in a 2019 examination of the effectiveness of the UK national DNA database. These indicators included (1) implementation cost – the financial input required to implement the database system, (2) crime-solving capability – the ability of the database to assist criminal justice officials in case resolution, (3) incapacitation effect – the ability of the database to reduce crime through the incapacitation of offenders, (4) deterrence effect – the preventative potential of the database through deterrence of individuals from committing crime, (5) privacy protection – protection of the privacy or civil liberty rights of individuals, (6) legitimacy – compliance of the databasing system to the principle of proportionality, and (7) implementation efficiency – the time and non-monetary resource required to implement the database system [ 79 ].
A follow-up article concluded: “Available evidence shows that while DNA analysis has contributed to successful investigations in many individual cases, its aggregate value to the resolution of all crime is low” [ 80 ]. The systematic review of 19 articles on DNA databases cited previously noted “the expansion of DNA databases would only have positive effects on detection and clearance if the offender were already included in the database” [ 63 ]. When previous offenders are not already in a law enforcement DNA database to provide a hit to a crime scene profile, efforts are increasingly turning to familial searching and investigative genetic genealogy as described in the following sections.
2.2.2. Familial DNA searching
Familial DNA searching (FDS) extends the traditional direct matching of STR profiles within law enforcement databases to search for potential close family relationships, such as a parent or sibling, of a profile in the database. 35 FDS typically uses Y-STR lineage testing to narrow the set of candidate possibilities along with other case information such as geographic details of the crime and age of the person(s) of interest. For example, FDS helped solve murder cases in Romania [ 81 ] and China [ 82 ] by locating the perpetrator through a relative in the DNA database. A survey of 103 crime laboratories in the United States found that 11 states use FDS while laboratories in 24 states use a similar but distinct practice of partial matching [ 83 ].
The expansion of the number of STRs from 15 to 20 or 21 helps distinguish between true and false matches during a DNA database search by reducing the number of FDS adventitious matches [ 84 ]. Another study noted that the choice of allele frequencies affects the rate at which non-relatives are erroneously classified as relatives and found that using ancestry inference on the query profile can reduce false positive rates [ 85 ]. New Y-STR kits have been developed to assist with familial searching [ 86 , 87 ]. FDS of law enforcement databases differs from investigative genetic genealogy in two important ways – the genetic markers and the databases used for searching [ 88 , 89 ].
2.2.3. Investigative genetic genealogy
In recent years when national DNA databases fail to generate a lead to a potential person of interest, law enforcement agencies have started to utilize the capabilities of investigative genetic genealogy (IGG), also called forensic genetic genealogy (FGG) or forensic investigative genetic genealogy (FIGG), as an approach to locate potential persons of interest in criminal or missing persons cases. For example, a pilot case study in Sweden used IGG to locate the perpetrator of a double murder from 2004 who had evaded detection despite 15 years of various investigation efforts including more than 9000 interrogations and mass DNA screenings of more than 6000 men [ 90 ]. Hardly a week goes by without mention in the global media of another cold case being solved with IGG. Since the arrest of Joseph DeAngelo in April 2018 identified as the infamous Golden State Killer using IGG, hundreds of cold criminal and unidentified human remains cases have been resolved [ 91 ].
IGG involves examination of about 600,000 single nucleotide polymorphisms (SNPs), rather than the 20 or so STRs used in conventional forensic DNA testing, to enable associations of relatives as distant as third or fourth cousins [ 17 ]. IGG relies on a combination of publicly accessible records and the consent of individuals who have uploaded their genetic genealogy DNA profiles to genetic genealogy databases [ 92 ]. Multiple reviews and research articles have been published describing current IGG methods, knowledge, and practice along with the effectiveness and operational limits of the technique [ 17 , 30 , [93] , [94] , [95] , [96] , [97] ]. IGG works best with high-quality, single-source DNA samples. A case study involving whole genome sequencing of human remains from a 2003 murder victim found that it was possible to perform IGG for identification of the victim in this situation [ 98 ].
The four main direct-to-consumer (DTC) genetic genealogy companies, 23andMe (Mountain View, CA), Ancestry (Salt Lake City, UT), FamilyTree DNA (Houston, TX), and My Heritage (Lehi, UT), have DNA data from over 41 million individuals 36 as of July 2022 [ 97 ]. Individuals can upload their DTC data to GEDmatch, which is a DNA comparison and analysis website launched in 2010 and purchased in 2019 by Verogen (San Diego, CA). Law enforcement IGG searches are currently permitted with DTC data for individuals who opt into the GEDmatch database or do not opt out of the FamilyTree DNA database [ 99 , 100 ]. Currently most DTC genetic genealogy data comes from the United States and individuals of European origin. A UK study found that 4 of 10 volunteer donors could be identified with IGG including someone of Indian heritage demonstrating that under the right circumstances individuals of non-European origin can be identified [ 101 ].
As noted previously in Section 1.2.1 , the U.S. Department of Justice released an interim policy guide to forensic genetic genealogical DNA analysis and searching [ 23 ], and the FBI Laboratory's chief biometric scientist published an editorial in Science calling for responsible genetic genealogy [ 24 ]. SWGDAM has provided an overview of IGG that emphasizes the approach being used only after a regular STR profile search of a law enforcement DNA database fails to produce any investigative leads [ 102 ]. Policy and practical implications of IGG have been explored in Australia [ 103 ] and within the UK as part of probing the perceptions of 45 professional and public stakeholders [ 104 , 105 ].
Four misconceptions about IGG were examined by several members of the SWGDAM group: (1) when law enforcement conducts IGG in a genetic genealogy database, they are given special access to participants' SNP profiles, (2) law enforcement will arrest a genetic genealogy database participant's relatives based on the genetic information the participant provided to the database, (3) IGG necessarily involves collecting and testing DNA samples from a larger number of innocent persons than would be the case if IGG were not used in the investigation, and (4) IGG is or soon will be ubiquitous because there are no barriers to IGG that limit the cases in which it can be conducted [ 106 ].
In May 2021, the state of Maryland passed the first law in the United States and in the world that regulates law enforcement's use of DTC genetic data to investigate crimes. A policy forum article in Science explained how this new law provides a model for others in this area [ 107 ]. Six important features were described: (1) requiring judicial authorization for the initiation of an IGG search, (2) affirming individual control over the investigative use of one's genetic data, (3) establishing strong protections for third parties who are not suspects in the case, (4) ensuring that IGG is available to prove either guilt or innocence, (5) imposing consequences and fines for violations, and (6) requiring annual public reporting and review to enable informed oversight of IGG methods. However, as of September 2022, these regulations have not been implemented apparently due to lack of resources with these unfunded requirements. 37
Efforts have been made to raise awareness among defense attorneys about how IGG searches can potentially invade people's privacy in unique ways [ 108 ]. Important perspectives on ethical, legal, and social issues have been offered along with directions for future research [ 109 ]. These concerns about data privacy, public trust, proficiency and agency trust, and accountability have led to a call for standards and certification of IGG to address issues raised by privacy scholars, law enforcement agencies, and traditional genealogists [ 110 , 111 ] and for an ethical and privacy assessment framework covering transparency, access criteria, quality assurance, and proportionality [ 112 ].
2.2.4. Genetic privacy and ethical concerns
Two important topics are considered in this section: (1) do the genetic markers used in traditional forensic DNA typing reveal more than identity and therefore potentially impact privacy of the individuals tested? and (2) are samples collected and tested according to ethical principles?
Forensic DNA databases utilize STR markers that were intentionally selected to avoid phenotypic associations. An extensive review of the literature examined 107 articles associating a forensic STR with some genetic trait and found “no demonstration of forensic STR variants directly causing or predicting disease” [ 113 ]. A study of the potential association of 15 STRs and 3 facial characteristics on 721 unrelated Han Chinese individuals also found “scarcely any association between [the] STRs with studied facial characteristics” [ 114 ].
In 2021, the American Type Culture Collection (ATCC) published a standard for authentication of human cell lines using DNA profiling with the 13 CODIS STR markers [ 115 ]. This use of forensic STR markers for biospecimen authentication led a bioethicist and a law professor to write a policy forum article in Science titled “Get law enforcement out of biospecimen authentication” [ 116 ]. The authors of this policy forum believe that using the same genetic markers could potentially: (1) undermine efforts to recruit research participants from historically marginalized and excluded groups that are underrepresented in research, (2) risk drawing law enforcement interest in gaining access to these research data, and (3) impose additional potential harms on already vulnerable populations, particularly children. Instead they advocate for using non-CODIS STRs or a new SNP assay to distinguish biospecimens in repositories, something done recently at the Coriell Institute for Medical Research with six new STR markers [ 117 ]. A responsive letter to the editor regarding this policy forum article expressed that “their proposal could potentially create artificial silos between genomic data in the justice system and in biomedical research, making it inefficient and ultimately counterproductive” [ 118 ]. The authors of the original article responded that “the risk of attracting law enforcement interest to research data increases when the data are available in a recognizable way” [ 119 ].
Modern scientific research seeks to protect the dignity, rights, and welfare of research participants by following ethical requirements. Six forensic science journals over the time period of 2010–2019 were examined for their reporting of ethical approval and informed consent in original research using human or animal subjects [ 120 ]. These journals were Forensic Science International: Genetics , Science & Justice , Journal of Forensic and Legal Medicine , the Australian Journal of Forensic Sciences , Forensic Science International , and the International Journal of Legal Medicine . A total of 3010 studies that described research on human or animal subjects and/or samples were selected from these journals with only 1079 articles (36%) reporting that they had obtained ethical approval and 527 articles (18%) stating that informed consent was sought either by written or verbal agreement. The authors of this study noted that reported compliance with ethical guidelines in forensic science research and publication was below what is considered minimal reporting rates in biomedical research and encouraged widespread adoption of the 2020 guidelines described below [ 120 ].
Guidelines and recommendations for ethnical research on genetics and genomics of biological material were jointly adopted and published in Forensic Science International: Genetics [ 121 ] and Forensic Science International: Reports [ 122 ]. These guidelines utilize the following principles as prerequisites for publication in these two journals as well as the Forensic Science International: Genetics Supplement Series : (1) general ethics principles that are regulated by national boards and represent widely signed international agreements, (2) universal declarations that require implementations in state members, such as the World Medical Association Declaration of Helsinki biomedical research on human subjects, and (3) universal declarations and principles drafted by independent organizations that have been widely adopted by the scientific community. This includes the U.S. Federal Policy for the Protection of Human Subjects (“Common Rule”) that was revised in 2017 (with a compliance date delayed to January 21, 2019). 38
Submitted manuscripts must provide the following supporting documentation to demonstrate compliance with the publication guidelines: (1) ethical approval in the country of [sample] collection by the appropriate local ethical committee or institutional review board, (2) ethical approval in the country of experimental work according to local legislation; if material collection and experimentation are conducted in different countries, both (1) and (2) are required, (3) template of consent forms in the case of human material as approved by the relevant ethical committee, and (4) approved export/import permits as applicable. Authors must declare in their submitted manuscript that these guidelines have been strictly followed [ 121 , 122 ].
Forensic genetic frequency databases, such as the Y-chromosome Haplotype Reference Database (YHRD), have been challenged over the ethics of DNA holdings, specifically of samples originating from the minority Muslim Uyghur population in western China [ 123 , 124 ]. A survey of U.S. state policies on potential law enforcement access to newborn screening samples found that nearly one-third of states permit these samples or their related data to be disclosed to or used by law enforcement and more than 25% of states have no discernible policy in place regarding law enforcement access [ 125 ].
A framework for ethical conduct of forensic scientists as “lived practice” has been proposed, and three case studies were discussed in terms of decision-making processes involving forensic DNA phenotyping and biographical ancestry testing, investigative genetic genealogy, and forensic epigenetics [ 126 ]. An ethos for forensic genetics involving the values of integrity, trustworthiness, and effectiveness has likewise been described [ 127 ].
2.2.5. Sexual assault kit testing
Unsubmitted or untested sexual assault kits (SAKs) may exist in police or laboratory evidence lockers for many years leading to rape kit backlogs that can spark community outrage when discovered. A number of articles have been published in the past three years describing success rates with examining SAKs and the policies surrounding them. For example, an evaluation of 3422 unsubmitted SAKs in Michigan found 1239 that produced a DNA profile eligible for upload into CODIS with 585 yielding a CODIS hit [ 128 ]. In addition, results from a groping and sexual assault case were presented to support the expansion of touch DNA evidence in these types of cases [ 129 ].
To assess success rates in their jurisdiction, the Houston Police Department randomly selected 491 cases of over 6500 previously unsubmitted sexual assault kits [ 130 ]. Of these, 336 cases (68%; 336/491) screened positive for biological evidence; a DNA profile was developed in 270 cases (55%; 270/491) with 213 (43%; 213/491) uploaded to CODIS; and 104 (21% total; 104/491 or 49% of uploaded profiles; 104/213) resulted in a CODIS hit. The statute of limitation had expired in 44% of these CODIS-hit cases, which prohibited arrests and prosecution. Victims were unwilling to participate in a follow-up investigation in another 25% of these cases. When the data were compiled for the publication, charges had been filed in only one CODIS-hit case [ 130 ].
Sexual assault cases can be difficult to prosecute as victims may be re-traumatized when a cold case is reopened. The authors of one study shared: “A key to successful pursuit of cold case sexual assaults is to have a well-crafted victim-notification plan and a victim advocate as part of the investigative team” [ 131 ]. Interviews with eight assistant district attorneys provided important prosecutors’ perspectives on SAK cases, the development of narratives to explain the evidence in a case, and the decision on whether a case should be pursued or what further investigative activities may be needed [ 132 ]. The authors concluded: “Our findings suggest that forensic evidence does not magically lead to criminal justice outcomes by itself, but must be used thoughtfully in conjunction with other evidence as part of a well-considered strategy of investigation and prosecution” [ 132 ].
Discussing a data set from Denver, Colorado where 1200 sexual assault cold cases with testable DNA samples were examined and 600 cases were processed through the laboratory resulting in 97 CODIS hits, 55 arrests and court filings, and 48 convictions, the authors conclude that the cost of the Denver cold case sexual assault program was worth the investment [ 131 ].
From December 2015 to July 2018, the Palm Beach County Sheriff's Office (Florida, USA) researched more than 5500 cases and evaluated evidence from previously untested sexual assault kits spanning a 43-year period at a cost of over $1 million. Of the 1558 sexual assaults examined, there were 686 cases (44%; 686/1558) with CODIS-eligible profiles, 261 CODIS hits, and 5 arrests when the article was written in mid-2019 [ 133 ]. The Palm Beach County Sheriff's Office also helped develop a backlog reduction effort through creating a biological processing laboratory within the Boca Raton Police Services Department [ 134 ]. With this joint effort from 2016 to 2018, the total average turnaround time decreased from 30 days to under 20 days with the 3489 DNA profiles entered into CODIS resulting in 1254 associations and 965 investigations aided. Important takeaway lessons include the value of (1) engaging legal counsel early to outline necessary legal procedures and the timeline, (2) bringing all stakeholders “to the table” early to discuss expectations, as well as legal and operational responsibilities, and (3) creating a realistic timeline with a comprehensive memorandum of understanding so all parties have agreed to their roles and responsibilities [ 134 ].
From 275 previously untested sexual assault kits submitted for DNA testing in one region of Central Brazil, a total of 176 profiles were uploaded to their DNA database resulting in 60 matches (34%; 60/176) and 32 assisted investigations (18%; 32/176) with information about the suspect identity or the connection of serial sexual assaults assigned to the same individual [ 135 ]. Another study from the same region of Brazil examined 2165 cases and noted that 13% (286/2165) had information regarding the victim-offender relationship with 63% (179/286) being stranger-perpetrated rapes and 37% (107/286) being non-stranger [ 136 ]. The authors then summarize: “Hits were detected only with stranger-perpetrated assaults ( n = 41), which reinforces that DNA databases are fundamental to investigate sexual crimes. Without DNA typing and DNA databases, probably these cases would never be solved” [ 136 ].
Given that laboratories have limited resources and need to prioritize their efforts, some business analytics have been applied to SAK testing. An analysis of the potential societal return on investment (ROI) for processing backlogged, untested SAKs reported a range of 10%–65% ROI depending on the volume of activity for the laboratory conducting the analysis [ 137 ]. An evaluation of data from 868 SAKs tested by the San Francisco Policy Department Criminalistics Laboratory during 2017–2019 found that machine learning algorithms outperformed forensic examiners in flagging potentially probative samples [ 138 ].
An examination of 5165 SAKs collected in Cuyahoga County (Ohio, USA) from 1993 through 2011 found 3099 with DNA of which 2127 produced a CODIS hit, with 803 investigations leading to an indictment and eventually 78 to trial along with 330 pleas [ 139 ]. The authors report a “cost savings to the community of $26.48 million after the inclusion of tangible and intangible costs of future sexual assaults averted through convictions” and advocate for “the cost-effectiveness of investigating no CODIS hit cases and support an ‘investigate all’ approach” [ 139 ]. Likewise an assessment of 900 previously-untested SAKs from Detroit (Michigan, USA) found that “few of the tested variables were significant predictors of CODIS hit rate” and “testing all previously-unsubmitted kits may generate information that is useful to the criminal justice system, while also potentially addressing the institutional betrayal victims experienced when their kits were ignored” [ 140 ].
A group in the Philippines described an integrated system to improve their SAK processing [ 141 ]. With an optimized workflow in Montreal, Canada, SAK processing median turnaround time decreased from 140 days to 45 days with a foreign DNA profile being obtained in 44% of cases [ 142 ]. In addition, this group examined casework data to guide resource allocation through identifying the likelihood of specific types of cases and samples yielding foreign biological material [ 142 ]. Decision trees and logistic regression models were also used to try and predict whether or not SAKs will yield a CODIS-eligible DNA profile [ 143 ]. Finally, direct PCR and rapid DNA approaches to streamline SAK testing were reviewed [ 144 ].
2.3. Forensic biology and body fluid identification
The basic workflow for biological samples in forensic examinations typically involves a visual examination of the evidence, a presumptive and/or confirmatory test for a suspected body fluid (e.g., the amylase assay for saliva), and DNA analysis and interpretation [ 145 ]. Body fluid identification (BFID), in particular with blood, saliva, semen, or vaginal fluid stains, provides valuable evidence in many investigations that can aid in the resolution of a crime [ 146 ]. Many of these BFID tests are presumptive and not nearly as sensitive as modern DNA tests meaning that “obtaining a DNA profile without being able to associate [it] with a body fluid is an increasingly regular occurrence” and “it is necessary and important, especially in the eyes of the law, to be able to say which body fluid that the DNA profile was obtained from” [ 147 ].
A number of approaches are being taken to improve the sensitivity and specificity of BFID in recent years including DNA methylation [ [148] , [149] , [150] , [151] , [152] , [153] , [154] , [155] , [156] , [157] , [158] , [159] , [160] , [161] ], messenger RNA (mRNA) [ [162] , [163] , [164] , [165] , [166] ], microRNA (miRNA) [ 167 ], protein mass spectrometry for seminal fluid detection [ 168 ], and microbiome analysis [ 169 , 170 ]. Although many new techniques are being described in the scientific literature, traditional methods for semen identification are still widely used in regular forensic casework [ 171 ].
When using RNA assays, DNA and RNA are co-extracted from examined samples [ 172 , 173 ]. Some tests may only distinguish between two possible body fluids, such as saliva and vaginal fluid [ 174 ], while other tests may attempt to distinguish six forensically relevant body fluids – vaginal fluid, seminal fluids, sperm cells, saliva, menstrual blood, and peripheral blood – although not always as clearly as desired [ 175 ]. BFID assays must also cope with mixed body fluids [ 176 ].
2.4. DNA collection and extraction
The process of obtaining a DNA profile begins with collecting a biological sample and extracting DNA from it. A review of recent trends and developments in forensic DNA extraction focused on isolating male DNA in sexual assault cases, using portable rapid DNA testing instruments, recovering DNA from difficult samples such as human remains, and bypassing DNA extraction altogether with direct PCR methods [ 177 ].
2.4.1. Touch evidence and fingerprint processing methods
Various studies have explored the compatibility of common fingerprint processing methods with DNA typing results [ [178] , [179] , [180] , [181] , [182] , [183] , [184] , [185] , [186] , [187] , [188] ]. For example, DNA recovery was explored after various steps in three different latent fingerprint processing methods – and fewer treatments were judged preferable with a 1,2-indanedione-zinc (IND/Zn) method appearing least harmful to downstream DNA analysis [ 187 ]. A different study found improved recovery of DNA from cigarette butts following latent fingerprint processing with 1,8-diazafluoren-9-one (DFO) compared to IND/Zn [ 179 ].
DNA losses were quantified with mock fingerprints deposited on four different surfaces to better understand DNA collection and extraction method performance [ 189 ]. The application of Diamond Dye has been shown to enable visualization of cells deposited on surfaces without interfering with subsequent PCR amplification and DNA typing [ [190] , [191] , [192] ].
It was possible to recover DNA profiles from clothing that someone touched for as little as 2 s [ 193 ]. DNA sampling success rates from car seats and steering wheels were studied [ 194 ] and recovery of DNA from vehicle surfaces using different swabs was explored [ 195 ]. In addition, the double-swab technique, where a wipe using a wet swab is followed by a wipe with a dry one, was revisited with an observation that for non-absorbing surfaces, the first web swab yielded 16 times more DNA than the second dry swab [ 196 ]. Swabs of cotton, flocked nylon, and foam reportedly provided equivalent DNA recoveries for smooth/non-absorbing surfaces, and an optimized swabbing technique involving the application of a 60-degree angle and rotating the swab during sampling improved DNA yields for cotton swabs [ 197 ].
2.4.2. Results from unfired and fired cartridge cases
Ammunition needs to be handled to load a weapon and thus DNA from the handler may be deposited onto the ammunition via touch [ 198 ]. Important progress has been made in recovering DNA from ammunition such as unfired cartridges or fired cartridge cases (FCCs) that may remain at a crime scene after a weapon has been fired. Trace quantities of DNA recovered from firearm or FCC surfaces has been used to try and link results to gun-related crimes.
A 2019 review of the literature regarding obtaining successful DNA results from ammunition examined collection techniques, extraction methodologies, and various amplification kits and conditions [ 199 ]. A direct PCR approach detected more STR alleles than methods using DNA extraction, and the authors noted that mixtures are commonly observed from gun surfaces, bullets, and cartridges in both controlled experimental conditions and from actual casework evidence and they encourage careful interpretation of these results [ 200 ]. The development of a crime scene FCC collector was combined with a new DNA recovery method that uses a rinse-and-swab technique [ 201 ].
Research studies and review articles have considered factors affecting DNA recovery from cartridge cases and the impact of metal surfaces on DNA recovery [ [202] , [203] , [204] , [205] , [206] , [207] , [208] , [209] ]. Recovery of mtDNA from unfired ammunition components has been assessed for sequence quality [ 210 ].
2.5. DNA typing
Following collection of DNA evidence and its extraction from biological samples, the typical typing process involves DNA quantitation, PCR amplification of STR markers, and STR typing using capillary electrophoresis. Direct PCR avoids the DNA extraction and quantitation steps, which can improve recovery of trace amounts of DNA [ 211 , 212 ]. Whole genome amplification prior to STR analysis has also been examined to aid recovery of degraded DNA [ 213 ] and to enable profiling of single sperm cells [ 214 ].
PCR amplification using STR typing kits can sometimes produce artifacts that impact DNA interpretation including missing (null) alleles [ 215 ], false tri-allelic patterns [ 216 ] or extra peaks when amplified in the presence of microbial DNA [ [217] , [218] , [219] ].
Applied Biosystems Genetic Analyzers have been the primary means of performing multi-colored capillary electrophoresis for many years [ 4 ]. First experiences with Promega's new Spectrum Compact CE System have recently been reported [ 220 ]. A number of new research and commercial STR kits have been introduced in recent years along with the publication of at least 24 validation studies ( Table 5 ). These validation studies typically follow guidelines outlined by the ENFSI DNA Working Group, 39 SWGDAM 40 , or a 2009 Chinese National Standard. 41
STR kits assessed with 24 published validation studies during 2019–2022.
| Publication | STR Kit/Primer Set | Comments |
|---|---|---|
| Al Janaahi et al., 2019 [ ] | VeriFiler Plus | Validation studies (sensitivity, peak height ratio, precision, reproducibility, thresholds, mixtures, concordance) |
| Alsafiah et al., 2019 [ ] | SureID 23comp Human Identification | Validation studies (following ENFSI and SWGDAM guidelines); has 17 non-CODIS STRs |
| Bai et al., 2019 [ ] | DNATyper25 | Validation studies (following SWGDAM and China National Standard); has 20 non-CODIS STRs |
| Cho et al., 2021 [ ] | Investigator 24plex QS, PowerPlex Fusion, GlobalFiler | Examined 189 casework samples and compared performance across the three kits |
| Fan et al., 2021 [ ] | STRtyper-32G | Developmental validation studies (SWGDAM); has 10 non-CODIS STRs |
| Green et al., 2021 [ ] | VeriFiler Plus | Developmental validation studies (SWGDAM); concordance checked with Huaxia Platinum kits |
| Hakim et al., 2020 [ ] | Investigator 24plex GO! | Validation studies; concordance with GlobalFiler |
| Harrel et al., 2021 [ ] | Investigator 24plex QS and GO! | Assessment of sample quality metrics in both kits |
| Jiang et al., 2021a [ ] | STRscan-17LC kit | Validation studies (SWGDAM) |
| Jiang et al., 2021b [ ] | Novel 8-dye STR multiplex | Validation studies (SWGDAM); 18 STRs plus AMEL; detection with GA118-24B Genetic Analyzer |
| Lenz et al., 2020 [ ] | VersaPlex 27PY system | Developmental validation studies (SWGDAM); includes D6S1043 |
| Li et al., 2021 [ ] | SureID S6 system | Validation studies (SWGDAM); concordance with Huaxia Platinum kit; uses lyophilized reagents |
| Liu et al., 2019 [ ] | 19 autosomal and 27 Y-STRs | Validation studies (Chinese National Standard); 47 loci (Fusion 6C, GlobalFiler, Yfiler Plus) with 6-dyes |
| Qu et al., 2019 [ ] | Microreader 20A ID system | Developmental validation studies (SWGDAM) |
| Qu et al., 2021 [ ] | Novel 6-dye, 31-plex | Developmental validation studies (SWGDAM and Chinese National Standard); 29 STRs, AMEL, Y-InDel |
| Wang et al., 2020a [ ] | 21plex with DYS391 and ABO | Describes a 21plex with 18 autosomal STRs, ABO blood group locus, DYS391, and AMEL |
| Wang et al., 2020b [ ] | Investigator 26plex QS kit | Validation studies (SWGDAM); concordance with AGCU Expressmarker 22 kit |
| Xie et al., 2020 [ ] | AGCU Expressmarker 16 + 22Y | Developmental validation studies (SWGDAM) |
| Xie et al., 2022 [ ] | Novel 26plex | Validation studies (SWGDAM); multiple STRs on chromosomes 13, 18, 21, and X for prenatal diagnosis |
| Yin et al., 2021 [ ] | Microreader 28A ID System | Developmental validation (SWGDAM); concordance with AGCU Expressmarker 22 kit |
| Zhang et al., 2020 [ ] | SiFaSTR 21plex_NCII | Developmental validation (SWGDAM); describes 18 new non-CODIS STR loci |
| Zhang et al., 2021 [ ] | AGCU Expressmarker 30 Kit | Developmental validation (SWGDAM); includes 6 non-CODIS STR loci; concordance with AGCU Expressmarker 22 kit |
| Zheng et al., 2019 [ ] | SiFaSTR 23-plex panel | Developmental validation (SWGDAM and Chinese National Standard) |
| Zhong et al., 2019 [ ] | Huaxia Platinum PCR kit | Developmental validation (SWGDAM and Chinese National Standards) |
A report on the first two years of submissions to the STRidER 42 (STRs for Identity ENFSI Reference) database for online allele frequencies revealed that 96% of the submitted 165 autosomal STR datasets generated by CE contained errors, showing the value of centralized quality control and data curation [ 245 ].
2.6. DNA interpretation at the source or sub-source level
The designation of STR alleles and genotypes of contributors in DNA mixtures are key aspects of DNA interpretation [ 246 , 247 ]. Electropherograms generated by CE instruments exhibit both STR alleles and artifacts that complicate data interpretation. Efforts are underway to understand and model instrumental artifacts [ [248] , [249] , [250] , [251] ] as well as biological artifacts of the PCR amplification process such as STR stutter products [ 252 , 253 ]. Machine learning approaches are being applied to classify artifacts versus alleles with the goal to eventually replace manual data interpretation with computer algorithms [ [254] , [255] , [256] , [257] ]. One such program, FaSTR DNA, enables potential artifact peaks from stutter, pull-up, and spikes to be filtered or flagged, and a developmental validation has been published examining 3403 profiles generated with seven different STR kits [ 258 ].
2.6.1. DNA mixture interpretation
Forensic evidence routinely contains contributions from multiple donors, which result in DNA mixtures. A number of approaches have been taken and advances made in DNA mixture interpretation [ 259 ]. These include probabilistic genotyping software [ 15 ], using genetic markers beyond traditional autosomal STR typing [ 260 ], or separating contributor cells and performing single-cell analysis [ [261] , [262] , [263] , [264] , [265] , [266] ].
In June 2021, the National Institute of Standards and Technology (NIST) released a draft report regarding the scientific foundations of DNA mixture interpretation [ 267 ]. This 250-page document described 16 principles that underpin DNA mixture interpretation, provided 25 key takeaways, and cited 528 references. NIST also began a Human Factors Expert Working Group on DNA Interpretation in February 2020 and plans to release a report with recommendations in 2023.
Assessment of the number of contributors (NoC) is a critical element of accurate DNA mixture interpretation. For example, the LRs relating to minor contributors can be reduced when the incorrect number of contributors is assumed [ 268 ]. Allele sharing among contributors to a mixture and masking of alleles due to STR stutter artifacts can lead to inaccurate NoC estimates based on simply counting the number of alleles at a locus. Different approaches and software programs have been used for NoC estimation [ [269] , [270] , [271] , [272] , [273] , [274] , [275] ]. Total allele count (TAC) distribution via TAC curves showed an improvement in manually estimating the number of contributors with complex mixtures [ 276 ]. Sequence analysis of STR loci expands the number of possible alleles compared to CE-based length measurements and thus can improve NoC estimates [ 277 ].
In the past three years, validation studies have been performed with a number of probabilistic genotyping software (PGS) systems including EuroForMix [ 278 ], DNAStatistX [ 279 , 280 ], TrueAllele [ 281 ], STRmix [ 282 ], Statistefix [ 283 ], Mixture Solution [ 284 ], Kongoh [ 285 ], and MaSTR [ 286 , 287 ]. Developers of EuroForMix, DNAStatistX, and STRmix provided a review of these systems [ 288 ]. Multi-laboratory assessments have been described [ 289 , 290 ] and likelihood ratios obtained from EuroForMix and STRmix compared [ [291] , [292] , [293] , [294] ]. With a growing literature in this area, there are many other articles that could have been cited.
2.7. DNA interpretation at the activity level
DNA interpretation at the source or sub-source level helps to answer the question of who deposited the cell material, whether attribution for the result can be made to a specific cell type (i.e., source level) or simply to the DNA if no attribution can be made to a specific cell type (i.e., sub-source level). Activity-level propositions seek to answer the question of how did an individual's cell material get there. Interpretation at the activity level is sometimes referred to as evaluative reporting [ 295 , 296 ].
In 2020, the ISFG DNA Commission [ 38 ] discussed the why, when, and how to carry out evaluative reporting given activity level propositions through providing examples of formulating these propositions. These Commission recommendations emphasize that reports using a likelihood ratio based on case-specific propositions and relevant conditioning information should highlight the assumptions being made and that “it is not valid to carry over a likelihood ratio from a low level, such as sub-source, to a higher level such as source or activity propositions … because the LRs given sub-source level propositions are often very high and LRs given activity level propositions will often be many orders of magnitude lower” [ 38 ]. Another recommendation specifies that “scientists must not give their opinion on what is the ‘most likely way of transfer’ (direct or indirect), as this would amount to giving an opinion on the activities and result in a prosecutor's fallacy (i.e., give the probability that X is true). The scientists' role is to assess the value of the results if each proposition is true in accordance with the likelihood ratio framework (the probability of the results if X is true and if Y is true)” [ 38 ] (emphasis in the original). This DNA Commission provided 11 recommendations and 4 considerations that should be studied carefully by those who implement activity-level DNA interpretation.
2.7.1. DNA transfer and persistence studies
To evaluate DNA findings given activity-level propositions it is important to understand the factors and variables that may impact DNA transfer, persistence, prevalence, and recovery (DNA-TPPR). These factors include history of contacting surfaces, biological material type, quantity and quality of DNA, dryness of biological material, manner and duration of contact, number and order of contacts, substrate type(s), time lapses and environment, and methods and thresholds used in the forensic DNA laboratory to generate the available data [ 297 ].
Three valuable review articles were published on this topic in 2019 [ 14 , 28 , 299 ]. Following a comprehensive January 2019 review that cited [ 298 ] references on DNA-TPPR [ 14 ], the same authors provided an update in November 2021 on recent progress towards meeting challenges and a synopsis of 144 relevant articles published between January 2018 and March 2021 [ 297 ]. While few studies provide the information needed to help assign probabilities of obtaining DNA results given specific sets of circumstances, progress includes use of Bayesian Networks [ 300 ] to identify variables for complex transfer scenarios [ 38 , [301] , [302] , [303] , [304] , [305] ] as well as development of an online database DNA-TrAC 43 for relevant research articles [ 299 ] and a structured knowledge base 44 with information to help practitioners interpret general transfer events at an activity level [ 306 ].
Forensic DNA pioneer Peter Gill emphasized that awareness of the limitations of DNA evidence is important for users of this data given that an increased sensitivity of modern DNA methods means that DNA may be recovered that is irrelevant to the crime under investigation [ 307 ]. An ISFG DNA Commission (see Section 1.2.5 ) emphasized that the strength of evidence associated with a DNA match at the sub-source level cannot be carried over to activity level propositions [ 38 ]. Structuring case details into propositions, assumptions, and undisputed case information has been encouraged [ 308 ].
Factors affecting variability of DNA recovery on firearms were studied with four realistic, casework-relevant handling scenarios along with results obtained including DNA quantities, number of contributors, and relative profile contributions for known and unknown contributors [ 309 ]. These studies found that sampling several smaller surfaces on a firearm and including the sampling location in the evaluation process can be helpful in assessing results given alternative activity-level propositions in gun-related crimes. The authors recommend that “further extensive, detailed and systematic DNA transfer studies are needed to acquire the knowledge required for reliable activity-level evaluations” [ 309 ].
Other recent studies on DNA-TPPR include examining prevalence and persistence of DNA or saliva from car drivers and passengers [ [310] , [311] , [312] ], evaluation of DNA from regularly-used knives after a brief use by someone else [ 313 ], studying the accumulation of endogenous and exogenous DNA on hands [ 314 ] and non-self-DNA on the neck [ 315 ], considering the potential of DNA transfer via work gloves [ 316 , 317 ] or during lock picking [ 318 ], and investigating whether DNA can be recovered from illicit drug capsules [ 319 , 320 ] or packaging [ 321 ] to identify those individuals preparing or handling the drugs.
Efforts have been made to estimate the quantity of DNA transferred in primary versus secondary transfer scenarios [ 322 ]. As quantities of DNA transferred can be highly variable and thought to be dependent on the so-called “shedder status” – how much DNA an individual exudes, several studies explored this topic [ [323] , [324] , [325] , [326] , [327] ]. Studies have also considered the level of DNA an individual transfers to untouched items in their immediate surroundings [ 328 ], the position and level of DNA transferred during digital sexual assault [ 329 ] or during various activities with worn upper garments [ 330 , 331 ], and the DNA composition on the surface of evidence bags pre- and post-exhibit examination [ 332 ]. Studies assessing background levels of male DNA on underpants worn by females [ 333 ] and background levels of DNA on flooring within houses [ 334 ] are providing important knowledge about the possibilities and probabilities of DNA transfer and persistence.
The authors of one study summarize some key points that could be extended to many other studies as words of caution: “From a wider trace DNA point of view, this study has demonstrated that the person who most recently handled an item may not be the major contributor and someone who handled an item for longer may still not be the major contributor if they remove more DNA than they deposit. The amount of DNA transferred and retained on an item is highly variable between individuals and even within the same individual between replicates” [ 320 ].
3. Emerging technologies, research studies, and other topics
New technologies to aid forensic DNA typing are constantly under development. This section explores recent activities with next-generation DNA sequencing, DNA phenotyping for estimating a sample donor's age, ancestry, and appearance, lineage markers, other markers and approaches, and non-human DNA and wildlife forensics, and is expected to be of value to researchers and those practitioners looking to future directions in the field.
3.1. Next-generation sequencing
Next-generation sequencing (NGS), also known as massively parallel sequencing (MPS) in the forensic DNA community, expands the measurement capabilities and information content of a DNA sample beyond the traditional length-based results with STR markers obtained with capillary electrophoresis (CE) methods. Additional genetic markers, such as single nucleotide polymorphisms (SNPs), microhaplotypes, and mitochondrial genome (mtGenome) sequence, may be analyzed along with the full sequence of STR alleles. This higher information content per sample opens up new potential applications such as phenotyping of externally visible characteristics and biogeographical ancestry as described in review articles [ 335 , 336 ].
As mentioned in Section 1.2.1 , the NIJ Forensic Laboratory Needs Technology Working Group (FLN-TWG) published a 29-page implementation strategy on next-generation sequencing for DNA analysis in September 2021 [ 28 ]. This guide discusses how NGS works and its advantages and disadvantages, the various instrument platforms and commercial kits available with approximate costs, items to consider regarding facilities, data storage, and personnel training, and resources for implementing NGS technology. A total of 73% of 105 forensic DNA laboratories surveyed from 32 European countries already own an MPS platform or plan to acquire one in the next year or two and one-third of the survey participants already conduct MPS-based STR sequencing, identity, or ancestry SNP typing [ 337 ].
Validation studies have been described with the ForenSeq DNA Signature Prep kit and the MiSeq FGx system [ [338] , [339] , [340] ], with the Verogen ForenSeq Primer Mix B for phenotyping and biogeographical ancestry predictions [ 341 , 342 ], and for resizing reaction volumes with the ForenSeq DNA Signature Prep kit library preparation [ 343 ]. MPS sequence data showed excellent allele concordance with CE results for 31 autosomal STRs in the Precision ID GlobalFiler NGS STR Panel from 496 Spanish individuals [ 344 ] and from 22 autosomal STR loci in the PowerSeq 46GY panel with 247 Austrians [ 345 ].
STR flanking region sequence variation has been explored [ 346 ] and reports of population data and sequence variation were published for samples from India [ 347 ], France [ 348 ], China [ 349 , 350 ], Korea [ 351 ], Brazil [ 352 ], Tibet [ 353 ], and the United States [ 354 ].
In April 2019 the STRAND ( S hort T andem R epeat: A lign, N ame, D efine) Working Group was formalized [ 355 ] to consider several possible approaches to sequence-based STR nomenclature that have been proposed [ 356 , 357 ]. An overview of software options has been provided for analysis of forensic sequencing data [ 358 ]. Some recent published options include STRinNGS [ 359 ], STRait Razor [ 360 ], ArmedXpert tools MixtureAce and Mixture Interpretation to analyze MPS-STR data [ 361 ], and STRsearch for targeted profiling of STRs in MPS data [ 362 ]. To aid interpretation of MPS-STR data, sensitivity studies were performed with single-source samples and sequence data analyzed by DNA quantity and method used [ 363 ]. A procedure has been described to address calculation of match probabilities when results are generated using MPS kits with different trim sites than those present in the relevant population frequency database [ 364 ]. Performance of different MPS kits, markers, or methods can be compared for accuracy and precision using the Levenshtein distance metric [ 365 ].
Novel MPS STR and SNP panels developed in recent years include IdPrism [ 366 ], a QIAGEN 140-locus SNP panel [ 367 ], the 21plex monSTR identity panel [ 368 ], a 42plex STR NGS panel to assist with kinship analysis [ 369 ], the 5422 marker FORCE (FORensic Capture Enrichment) panel [ 370 ], a forensic panel with 186 SNPs and 123 STRs [ 371 ], the SifaMPS panel for targeting 87 STRs and 294 SNPs [ 372 ], a 1245 SNP panel [ 373 ], 90 STRs and 100 SNPs for application with kinship cases [ 374 ], an adaption of the SNPforID 52plex panel to MPS [ 375 ], 448plex SNP panel [ 376 ], a 133plex panel with 52 autosomal and 81 Y-chromosome STRs [ 377 ], and a forensic identification multiplex with 1270 tri-allelic SNPs involving 1241 autosomal and 29 X-chromosome markers [ 378 ]. The 124 SNPs in the Precision ID Identity Panel were examined in a central Indian population [ 379 ] and human leukocyte antigen (HLA) alleles used in the early 1990s were revisited with MPS capability [ [380] , [381] , [382] ].
MPS methods have demonstrated utility with compromised samples [ [383] , [384] , [385] , [386] , [387] , [388] ] and mixture interpretation [ [389] , [390] , [391] , [392] , [393] , [394] , [395] ]. Microhaplotype assays have also been developed to assist with DNA mixture deconvolution [ 396 , 397 ]. Collaborative studies have explored variability with laboratory performance using MPS methods [ 398 , 399 ]. Population structure [ 400 ] and linkage and linkage disequilibrium [ 401 ] were examined among the markers in forensic MPS panels.
A review of transcriptome analysis using MPS discussed efforts with body fluid and tissue identification, determination of the time since deposition of stains and the age of donors, the estimation of post-mortem interval, and assistance to post-mortem death investigations [ 402 ]. The potential for MPS methods to assist with environmental trace analysis was reviewed in terms of forensic soil analysis, forensic botany, and human identification utilizing the skin microbiome [ 403 ]. The possibility of non-invasive prenatal paternity testing using cell-free fetal DNA from maternal plasma was explored with the Precision ID Identity Panel [ 404 ] and the ForenSeq DNA Signature Prep Kit [ 405 ]. Pairwise kinship analysis was also examined using the ForenSeq DNA Signature Prep Kit and multi-generational family pedigrees [ 406 , 407 ]. Nanopore sequencing has also been explored for sequencing STR and SNP markers [ [408] , [409] , [410] , [411] , [412] , [413] , [414] , [415] , [416] ].
3.2. DNA phenotyping (ancestry, appearance, age)
Continuing research into the genetic components of biogeographic ancestry, appearance, and age predictions have improved forensic DNA phenotyping capabilities [ 417 ]. These forensic innovations may sometimes impact public expectations [ 418 ]. The investigation in a murder case was assisted using information from forensic DNA phenotyping that predicted eye, hair, and skin color of an unknown suspect with the HIrisPlex-S system involving targeted massively parallel sequencing [ 419 ].
The VISAGE ( Vis ible A ttributes Through Ge nomics) Consortium, which consists of 13 partners from academic, police, and justice institutions in 8 European countries, has established new scientific knowledge and developed and tested prototype tools for DNA analysis and statistical interpretation as well as conducted education for stakeholders. In the 2019 to 2022 time window of this review, this concerted effort produced 45 one review article [ 417 ], 22 original research publications [ 337 , [420] , [421] , [422] , [423] , [424] , [425] , [426] , [427] , [428] , [429] , [430] , [431] , [432] , [433] , [434] , [435] , [436] , [437] , [438] , [439] , [440] ], and three reports [ [441] , [442] , [443] ].
DNA phenotyping is currently an active area of research, and numerous activities and publications exist beyond the VISAGE articles noted here. Another 137 articles have appeared in the literature in the past three years on biogeographical ancestry, appearance (primarily hair color, eye color, and skin color), and biological age predictions (typically utilizing DNA methylation) (see Supplemental File ).
3.3. Lineage markers (Y-chromosome, mtDNA, X-chromosome)
Lineage markers consist of Y-chromosome, mitochondrial DNA, and X-chromosome genetic information that may be inherited from just one parent without the regular recombination that occurs with autosomal DNA markers. Research in terms of new markers, assays, and population studies continue to be published for these lineage markers.
3.3.1. Y-chromosome
Several recent review articles were published on forensic applications of Y-chromosome testing [ [444] , [445] , [446] ]. As discussed previously in Section 1.2 , an ISFG DNA Commission summarized the state of the field with Y-STR interpretation [ 39 ]. Rapidly mutating Y-STR loci can be used to differentiate closely related males [ [447] , [448] , [449] ]. New statistical approaches to assessing evidence with Y-chromosome information have been described [ 450 , 451 ]. Four commercial Y-STR multiplexes were compared with the NIST 1032 U S. population sample set and the allele and haplotype diversities explored with length-based versus sequence-based information [ 452 ].
A number of Y-STR typing systems have been described along with validation studies, such as a 36plex [ 453 ], a 41plex [ 454 ], a 29plex [ 455 ], a 17plex [ 456 ], a 24plex [ 457 ], the Microreader 40Y ID System [ 458 ], the 24 Y-STRs in the AGCU Y SUPP STR kit [ 459 ], the DNATyper Y26 PCR amplification kit [ 460 ], a multiplex with 12 multicopy Y-STR loci [ 461 ], the Yfiler Platinum PCR Amplification Kit [ 462 ], a 45plex [ 463 ], the Microreader 29Y Prime ID system [ 464 ], an assay with 30 slow and moderate mutation Y-STR markers [ 465 ], the 17plex Microreader RM-Y ID System [ 466 ], and a 26plex for rapidly mutating Y-STRs [ 467 ]. A machine learning program predicted Y haplogroups using two Y-STR multiplexes with 32 Y-STRs [ 468 ].
Deletions and duplications with 42 Y-STR were reported in a sample of 1420 unrelated males and 1160 father-son pairs from a Chinese Han population [ 469 ]. Using Y-STR allele sequences has enabled locating parallel mutations in deep-rooting family pedigrees [ 470 ]. The surname match frequency with Y-chromosome haplotypes was explored using 2401 males genotyped for 46 Y-STRs and 183 Y-SNPs [ 471 ]. In the Y-chromosome's role as a valuable kinship indicator to assist in genetic genealogy and forensic research, models to improve prediction of the time to the most recent common paternal ancestor have been studied with 46 Y-STRs and 1120 biologically related genealogical pairs [ 472 ]. A massively parallel sequencing tool was developed to analyze 859 Y-SNPs to infer 640 Y haplogroups [ 473 ]. Another MPS tool, the CSYseq panel, targeted 15,611 Y-SNPs to categorize 1443 Y-sub-haplogroup lineages worldwide along with 202 Y-STRs including 81 slow, 68 moderate, 27 fast, and 26 rapidly mutating Y-STRs to individualize close paternal relatives [ 474 ].
3.3.2. Mitochondrial DNA
Mitochondrial DNA (mtDNA), which is maternally inherited with a high copy number per cell, can aid human identification, missing persons investigations, and challenging forensic specimens containing low quantities of nuclear DNA such as hair shafts [ [475] , [476] , [477] ]. Validation studies have been published using traditional Sanger sequencing [ 478 ] and next-generation sequencing [ [479] , [480] , [481] ]. Illumina and Thermo Fisher now provide mtDNA whole genome NGS assays [ [482] , [483] , [484] , [485] ]. Many mtDNA population data sets were published in the past three years including high-quality data from U.S. populations [ 486 ]. The suitability of current mtDNA interpretation guidelines for whole mtDNA genome (mtGenome) comparisons has been evaluated [ 487 ].
NGS methods have increased sensitivity of mtDNA heteroplasmy detection [ 488 , 489 ], which can influence the ability to connect buccal reference samples and rootless hairs from the same individual [ 490 , 491 ]. Twelve polymerases were compared in terms of mtDNA amplification yields from challenging hairs – with KAPA HiFi HotStart and PrimeSTR HS outperforming AmpliTaq Gold DNA polymerase that is widely used in forensic laboratories [ 492 ]. Multiple studies and review articles have discussed distinguishing mtDNA from nuclear DNA elements of mtDNA (NUMTs) that have been inserted into our nuclear DNA [ [493] , [494] , [495] , [496] ].
NGS sequencing of the mtGenome has permitted improved resolution of the most common West Eurasian mtDNA control region haplotype [ 497 ]. Phylogenetic alignment and haplogroup classification have continued to be refined with new sequence information [ 498 ], and new assays have been developed to aid haplogroup classification [ 499 ]. Concerns over potential paternal inheritance of mtDNA have also been addressed [ 500 , 501 ].
3.3.3. X-chromosome
A 20-year review of X-chromosome use in forensic genetics examined the number and types of markers available, an overview of worldwide population data, the use of X-chromosome markers in complex kinship testing, mutation studies, current weaknesses, and future prospects [ 502 ]. One example of the forensic application of X-chromosome markers include use in relationship testing cases involving suspicion of incest or paternity without a maternal sample for comparison [ 503 ]. Four new X-STR multiplex assays were described along with validation studies including a 19plex [ 504 ], a 16plex [ 505 ], another 19plex – the Microreader 19X Direct ID System [ 506 ], and an 18plex named TYPER-X19 multiplex assay [ 507 ]. A collaborative study examined paternal and maternal mutations in X-STR markers [ 508 ]. A software program for performing population statistics on X-STR data was introduced [ 509 ] and sequence-based U.S. population data described for 7 X-STR loci [ 510 ].
3.4. New markers and approaches (microhaplotypes, InDels, proteomics, human microbiome)
In this section on new markers and approaches, publications related to microhaplotypes and insertion/deletion (InDel, or DIP for deletion insertion polymorphisms) markers are reviewed along with proteomic and microbiome approaches to supplement standard human DNA typing methods.
3.4.1. Microhaplotypes
Microhaplotype (MH) markers consist of multiple SNPs in close proximity (e.g., typically <200 bp or <300 bp) that can be simultaneously genotyped with each DNA sequence read using NGS. Two or more linked SNPs will define three or more haplotypes. Compared to STR markers, MHs do not have stutter artifacts (which complicate mixture interpretation), can be designed with shorter amplicon lengths in some cases (which benefits recovery of genetic information from degraded DNA samples), possess a higher degree of polymorphism compared to single SNP loci (which benefits discrimination power), and exhibit low mutation rates (which enables relationship testing and biogeographical ancestry inference). Thus, MH markers bring advantages to human identification, ancestry inference, kinship analysis, and mixture deconvolution to potentially assist missing person investigations, relationship testing, and forensic casework as discussed in several recent reviews [ 16 , 511 ]. A new database, MicroHapDB, has compiled information on over 400 published MH markers and frequency data from 26 global population groups [ 512 ].
A number of MH panels have been described [ [513] , [514] , [515] , [516] , [517] , [518] , [519] ]. Population data has been collected from a number of sources around the world including four U.S. population groups examined with a 74plex assay with 74 MH loci and 230 SNPs [ 520 ]. Various MH panels have been evaluated for effectiveness with kinship analysis [ [521] , [522] , [523] ]. Likewise the ability to detect minor contributors in DNA mixtures has been assessed [ [524] , [525] , [526] ].
3.4.2. InDel markers
InDel markers can be detected using a CE-based length analysis, and thus use instrumentation that forensic DNA laboratories already have. InDels can also be designed to amplify short DNA fragments (e.g., <125 bp) to help improve amplification success rates with low DNA quantity and/or quality. However, with only two possible alleles like SNPs, InDels are not as polymorphic as STRs and thus require more markers to obtain similar powers of discrimination as multi-allelic STR markers and do not work as well with mixed DNA samples. InDels possess a lower mutation rate than STRs and can be used as ancestry informative markers (AIMs) since allele frequencies may differ among geographically separated population groups.
Two commercial InDel kit exist: (1) Investigator DIPlex (QIAGEN, Hilden, Germany) with 30 InDels [ [527] , [528] , [529] , [530] , [531] ] and (2) InnoTyper 21 (InnoGenomics, New Orleans, Louisiana, USA) with 21 autosomal insertion-null (INNUL) markers [ [532] , [533] , [534] , [535] ]. In addition, a number of InDel assays have been published including a 32plex [ 536 ], a 35plex [ 537 ], a 38plex [ 538 ], a 39plex with AIMs [ 539 ], a 43plex [ 540 ], a 57plex [ 541 ], a 60plex with 57 autosomal InDels, 2 Y-chromosome InDels, and amelogenin [ 542 ], a 32plex with X-chromosome InDels [ 543 ], and a 21plex with AIMs [ 544 ].
A multi-InDel marker is a specific DNA fragment with more than one InDel marker located tightly in the physical position that provides a microhaplotype [ 545 ]. Several multi-InDel assays have been published include a 12plex [ 546 ] and an 18plex [ 547 ].
3.4.3. Proteomics
Protein analysis, often through immunological assays, has traditionally been used to identify body fluids and tissues. With improvements in protein mass spectrometry in recent years, genetic variation can be observed in hair shafts via single amino acid polymorphisms. Detection of these genetically variant peptides (GVPs) can infer the presence of corresponding SNP alleles in the genome of the individual who is the source of the protein sample. A thorough review of forensic proteomics in 2021 cited 375 references [ 18 ]. Recent efforts in this area have focused on using GVPs to differentiate individuals through their human skin cells [ [548] , [549] , [550] ] or hair samples [ [551] , [552] , [553] , [554] , [555] , [556] , [557] , [558] , [559] ]. An algorithm has been proposed for calculating random match probabilities with GVP information [ 560 ].
3.4.4. Human microbiome
Microorganisms live in and on the human body, and efforts are underway to utilize the human microbiome for a variety of potential forensic applications [ 21 , [561] , [562] , [563] ]. There are also active efforts with analysis of microbiomes in the environment (e.g., soil or water samples), which could be classified under non-human DNA testing. Forensic microbiome research covers at least six areas: (1) individual identification, (2) tissue/body fluid identification, (3) geolocation, (4) time since stain deposition estimation, (5) forensic medicine, and (6) post-mortem interval (PMI) estimation. Biological, technical, and data issues have been raised and potential solutions explored in a recent review article [ 21 ]. For example, microbes on deceased individuals are being studied to estimate the postmortem interval [ 20 ] and postmortem skin microbiomes were found to be stable during repeated sampling up to 60 h postmortem [ 564 ].
Sequence analysis of 16S rRNA using NGS provides information on the microbiome community present in a tested sample [ 565 ]. The Forensic Microbiome Database 46 correlates publicly available 16S rRNA sequence data as a community resource. If the skin microbiome is extremely diverse among individuals, then the potential exists to associate the bacterial communities on an individual's skin with objects touched by this individual assuming that the bacteria originating from the donor's skin are deposited (i.e., transfer to and persist on the surface) and can be detected and interpreted.
Specific aspects of the microbiome (e.g., the bacterial community) may be able to provide details about the donor through bacterial profiling. For example, in one study correlations were observed between the bacterial profile and gender, ethnicity, diet type, and hand sanitizer used [ 566 ]. Another study with 30 individuals found that each person left behind microbial signatures that could be used to track interaction with various surfaces within a building, but the authors concluded “we believe the human microbiome, while having some potential value as a trace evidence marker for forensic analysis, is currently under-developed and unable to provide the level of security, specificity and accuracy required for a forensic tool” [ 565 ].
Direct and indirect transfer of microbiomes between individuals has been studied [ 567 , 568 ] along with identifying background microbiomes [ 569 ] and the possibility of transfer of microbiomes within a forensic laboratory setting [ 570 ]. Changes in four bacterial species in saliva stains were charted, showing that it was possible to correctly predict deposition time within one week in 80% of the stains [ 571 ]. The ability to detect sexual contact has been explored through using the microbiome of the pubic region [ [572] , [573] , [574] ]. The microbiomes on skin, saliva, vaginal fluid, and stool samples have been compared [ 575 ]. The stability, diversity, and individualization of the human skin virome was explored with 59 viral biomarkers being found that differed across the 42 individuals studied [ 576 ]. It will be interesting to see what the future holds and what other findings come from this active area of research.
3.5. Kinship analysis, human identification, and disaster victim identification
Kinship analysis, which uses genetic markers and statistics to evaluate the potential for specific biological relationships, is important for parentage testing, disaster victim identification (DVI), and human identification of remains that may be recovered in missing person cases. New open-source software programs have been described that can assist with kinship analysis [ 577 , 578 ].
A potential biological relationship is commonly evaluated using a likelihood ratio (LR) by comparing the likelihoods of observing the genetic data given two alternative hypotheses, such as (1) an individual is related to another individual in a defined relationship versus (2) the two individuals not related. Higher LR values indicate stronger support with the genetic data if the proposed relationship is true. Multiple factors influence LR kinship calculations including the specific hypotheses, the genetic markers examined, the allele frequencies of the relevant population(s), the co-ancestry coefficient applied, and approaches to address potential mutations. STR genotypes were reported for 11 population groups used by the FBI Laboratory [ 579 ]. The status quo has been challenged in recent articles regarding how hypotheses are commonly established [ 580 ] and whether race-specific U.S. population databases should be used for allele frequency calculations [ 581 ].
Depending on the relationship being explored, information can be optimized through genetic information from additional known relatives or through collecting results at more loci [ 582 ]. Potential error rates have been modeled with the observation that false negatives, which occur when related individuals are misinterpreted as being unrelated, are more common than false positives, where unrelated people are interpreted as being related [ 583 ]. While LRs are generally reliable in detecting or confirming parent/child pairs, limitations of kinship determinations exist (e.g., distinguishing siblings from half-siblings) when using STR data [ 584 ].
Pairwise comparisons have been studied in forensic kinship analysis [ [585] , [586] , [587] ]. The effectiveness of 40 STRs plus 91 SNPs was shown to be better than 27 STRs and 91 SNPs or 40 STRs alone [ 588 ]. Only a minor increase in LRs was observed when taking NGS-generated allele sequence variation rather than fragment length allele variation [ 589 ]. The statistical power of exclusion and inclusion can be used to prioritize family members selected for testing in resolving missing person cases [ 590 ]. A strategy for making decisions when facing low statistical power in missing person and DVI cases was published [ 591 ].
The most challenging kinship cases involve efforts to separate pairs of individuals who are typically thought to be genetically indistinguishable (i.e., monozygotic twins) or distant relatives (e.g., fourth cousins) where there is an increased uncertainty in the possible relationship. In some situations, somatic mutations may permit distinguishing monozygotic twins following whole genome sequencing – and this approach was successful in four of six cases reported recently [ 19 ]. The probative value of NGS data for distinguishing monozygotic twins was explored [ 592 ]. A unique case of heteropaternal twinning was reported where opposite-sex twins apparently had different fathers [ 593 ]. An impressive effort in kinship analysis using direct-to-consumer genetic genealogy information from 56 living descendants of multiple genealogical lineages helped resolve a contested paternity case from over a century and a half ago to identify the biological father of Josephine Lyon [ 594 ].
Techniques for identification of human remains continue to improve particularly with the capabilities of NGS and hybridization capture [ 595 ] and ancient DNA extraction protocols [ 596 , 597 ]. Studies have reported variation in skeletal DNA preservation [ 598 ] and retrospectively considered success rates with compromised human remains [ 599 ].
A simulated airplane crash enabled six forensic laboratories in Switzerland to gain valuable DVI experience with kinship cases of varying complexity [ 600 ]. The ISFG Spanish-Portuguese Speaking Working Group likewise conducted a DVI collaborative exercise with a simulated airplane crash to explore fragment re-associations, victim identification through kinship analysis, coping with related victims, handling mutations or insufficient number of family references, working in a Bayesian framework, and the correct use of DVI software [ 601 ]. Other groups have explored the capability of a particular software tool [ 602 ] or implemented rapid DNA analysis to accelerate victim identification [ 603 ]. The International Commission on Missing Persons (ICMP) has gained considerable experience with DNA extraction and STR amplification from degraded skeletal remains and kinship matching procedures in large databases [ 604 ]. To supplement the INTERPOL DVI Guide, 47 some lessons learned and experienced-based recommendations for DVI operations have recently been provided [ 605 ].
3.6. Non-human DNA testing and wildlife forensics
Non-human biological evidence may inform criminal investigations when animals or plants are victims or perpetrators of crime or the presence of specific material, such as cat or dog hair, may contribute to reconstructing events at a crime scene. Non-human DNA testing includes wildlife forensics and domestic animal species as well as forensic botany and has many commonalities and some important differences compared to human DNA testing [ [606] , [607] , [608] , [609] , [610] ]. Pollen analysis can assist criminal investigations [ 611 , 612 ]. The potential for and the barriers associated with the wider application of forensic botany in civil proceedings and criminal cases have been examined [ 613 , 614 ].
Mammalian species identification can assist in determining the origins of non-human biological material found at crime scenes through narrowing the range of possibilities [ 615 ]. New sequencing methods have been developed to assist species identification [ 616 ]. A multiplex PCR assay was developed to simultaneously identify 22 mammalian species (alpaca, Asiatic black bear, Bactrian camel, brown rat, cat, cow, common raccoon, dog, European rabbit, goat, horse, house mouse, human, Japanese badger, Japanese wild boar, masked palm civet, pig, raccoon dog, red fox, sheep, Siberian weasel, and sika deer) and four poultry species (chicken, domestic turkey, Japanese quail, and mallard) [ 617 ]. A number of other species identification assays have also been reported [ [618] , [619] , [620] ].
An important effort for harmonizing canine DNA analysis is an ISFG working group known as the Canine DNA Profiling Group, or CaDNAP. 48 The CaDNAP group published an analysis of 13 STR markers in 1184 dogs from Germany, Austria, and Switzerland [ 621 ]. Six traits for predicting visible characteristics in dogs, namely coat color, coat pattern, coat structure, body size, ear shape, and tail length, were explored with 15 SNPs and six InDel markers [ 622 ]. Canine breed classification and skeletal phenotype prediction has been explored using various genetic markers [ 623 ]. A novel assay using a feline leukemia virus was developed to demonstrate that a contested bobcat was not a domestic cat hybrid [ 624 ] and a core panel of 101 SNP markers was selected for domestic cat parentage verification and identification [ 625 ].
DNA tests have been developed to assist with illegal trafficking investigations involving elephant ivory seizures [ 626 ], falcons [ 627 ], and precious coral material [ 628 ]. Accuracy in animal forensic genetic testing was explored with interlaboratory assessments performed in 2016 and 2018 [ 629 ]. A collaborative exercise conducted in 2020 and 2021 by the ISFG Italian Speaking Working Group examined performance across 21 laboratories with a 13-locus STR marker test for Cannabis sativa [ 630 ]. A molecular approach was explored to distinguish drug-type versus fiber-type hemp varieties [ 631 ].
Acknowledgments and disclaimer
I am grateful to Dominique Saint-Dizier from the French National Scientific Police for the invitation and opportunity to conduct this review and for the support of my supervisor, Shyam Sunder, for granting the time to work on this extensive review. Input and suggestions on this manuscript by Todd Bille, Thomas Callaghan, Kevin Kiesler, François-Xavier Laurent, Robert Ramotowski, Kathy Sharpless, and Robert Thompson are greatly appreciated. Certain commercial entities, equipment, or materials may be identified in this document in order to describe an experimental procedure or concept adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the entities, materials, or equipment are necessarily the best available for the purpose.
1 https://www.sciencedirect.com/journal/forensic-science-international-genetics/special-issue/10TSDS4360H .
2 https://www.mdpi.com/journal/genes/special_issues/Forensic_Genetic .
3 https://www.mdpi.com/journal/genes/special_issues/forensic_mitochondrial_genomics .
4 https://www.mdpi.com/journal/genes/special_issues/Advances_Forensic_Genetics .
5 https://www.mdpi.com/books/pdfdownload/book/5798 .
6 https://www.mdpi.com/journal/genes/special_issues/Bioinformatics_Forensic_Genetics .
7 https://www.mdpi.com/journal/genes/special_issues/genetics_anthropology .
8 https://www.mdpi.com/journal/genes/special_issues/Identification_of_Human_Remains .
9 https://www.mdpi.com/journal/genes/special_issues/Forensic_DNA_analysis .
10 https://www.mdpi.com/journal/genes/special_issues/Forensic_DNA_Mixture .
11 https://www.mdpi.com/journal/genes/special_issues/28FBA0G4DH .
12 See https://www.swgdam.org/ .
13 https://www.swgdam.org/publications .
14 https://www.fbi.gov/file-repository/rapid-dna-guide-january-2022.pdf/view .
15 https://www.fbi.gov/file-repository/non-codis-rapid-dna-best-practices-092419.pdf/view .
16 https://www.fbi.gov/file-repository/rapid-dna-testing-for-non-codis-uses-considerations-for-court-073120.pdf/view .
17 https://www.justice.gov/olp/uniform-language-testimony-and-reports .
18 https://forensiccoe.org/human_factors_forensic_science_sourcebook/ .
19 https://www.nist.gov/organization-scientific-area-committees-forensic-science .
20 https://www.nist.gov/organization-scientific-area-committees-forensic-science/human-forensic-biology-subcommittee .
21 https://www.nist.gov/topics/organization-scientific-area-committees-forensic-science/wildlife-forensics-subcommittee .
22 https://www.aafs.org/academy-standards-board .
23 https://www.nist.gov/organization-scientific-area-committees-forensic-science/osac-registry .
24 See https://www.nist.gov/organization-scientific-area-committees-forensic-science/human-forensic-biology-subcommittee .
25 https://lexicon.forensicosac.org/ .
26 https://www.nist.gov/osac/human-factors-validation-and-performance-testing-forensic-science .
27 https://www.nist.gov/organization-scientific-area-committees-forensic-science/osac-research-and-development-needs .
28 https://www.gov.uk/government/publications/forensic-science-providers-codes-of-practice-and-conduct-2021-issue-7 .
29 https://www.aabb.org/standards-accreditation/standards/relationship-testing-laboratories .
30 https://www.isfg.org/DNA+Commission .
31 Previously available rapid DNA systems included the RapidHIT 200 from IntegenX and MiDAS (Miniaturized integrated DNA Analysis System) from the Center for Applied NanoBioscience at the University of Arizona.
32 See https://le.fbi.gov/science-and-lab-resources/biometrics-and-fingerprints/codis/rapid-dna .
33 See https://www.interpol.int/How-we-work/Forensics/DNA .
34 See https://www.interpol.int/How-we-work/Forensics/I-Familia .
35 See https://le.fbi.gov/science-and-lab-resources/biometrics-and-fingerprints/codis#Familial-Searching .
36 See https://isogg.org/wiki/Autosomal_DNA_testing_comparison_chart .
37 See https://www.wmar2news.com/infocus/maryland-quietly-shelves-parts-of-genealogy-privacy-law .
38 See https://www.hhs.gov/ohrp/regulations-and-policy/regulations/finalized-revisions-common-rule/index.html .
39 See https://enfsi.eu/about-enfsi/structure/working-groups/dna/ .
40 See https://www.swgdam.org/publications .
41 See https://www.chinesestandard.net/PDF/English.aspx/GAT815-2009 .
42 See https://strider.online/ .
43 See https://bit.ly/2R4bFgL (DNA-TrAC).
44 See https://cieqfmweb.uqtr.ca/fmi/webd/OD_CIEQ_CRIMINALISTIQUE (Transfer Traces Activity DataBase).
45 See https://www.visage-h2020.eu/index.html#publications .
46 See http://fmd.jcvi.org/ .
47 See https://www.interpol.int/en/How-we-work/Forensics/Disaster-Victim-Identification-DVI .
48 See https://www.isfg.org/Working+Groups/CaDNAP .
Appendix A Supplementary data to this article can be found online at https://doi.org/10.1016/j.fsisyn.2022.100311 .
Appendix A. Supplementary data
The following is the supplementary data to this article:
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 27 September 2024
Intelligent vehicle lateral control strategy research based on feedforward + predictive LQR algorithm with GA optimisation and PID compensation
- Zhu-an Zheng ORCID: orcid.org/0000-0002-1959-9119 1 ,
- Zimo Ye ORCID: orcid.org/0009-0003-7745-1002 1 &
- Xiangyu Zheng ORCID: orcid.org/0009-0000-3105-0885 1
Scientific Reports volume 14 , Article number: 22317 ( 2024 ) Cite this article
22 Accesses
Metrics details
- Engineering
- Mechanical engineering
Targeting the lateral motion control problem in the intelligent vehicle autopilot structural system, this paper proposes a feedforward + predictive LQR algorithm for lateral motion control based on Genetic Algorithm (GA) parameter optimisation and PID steering angle compensation. Firstly, based on the vehicle dynamics tracking error model, the intelligent vehicle LQR lateral motion controller as well as the feedforward controller are designed, and upon which the predictive controller is added to eliminate the system lag.Subsequently, exploiting the advantage that the PID algorithm is not model-based, a PID steering angle compensation controller that can directly control and correct the lateral error is designed. Second, a LQR controller based on path tracking deviation is designed by using the parameter rectification method of genetic algorithm (GA), which optimizes the control parameters of the lateral motion controller and improves the adaptivity of the control accuracy. Finally, Based on the Carsim-Simulink co-simulation platform, the simulation validation and analysis of double lane change (DLC) test and circular condition test (CCT) are carried out, and the results indicate that compared with the other two LQR controllers, the optimised controllers improved more than 50% in lateral error and heading error control, and the vehicle sideslip angle and vehicle yaw rate are in the range of −0.05° to 0.05° and − 0.15 rad/s to 0.10 rad/s, and it showed improved performance in tracking accuracy and satisfied vehicle stability constrains.
Similar content being viewed by others
Dynamic-boundary-based lateral motion synergistic control of distributed drive autonomous vehicle
Integrated control of braking-yaw-roll stability under steering-braking conditions
Distributed MPC of vehicle platoons with guaranteed consensus and string stability
Introduction.
Intelligent Vehicle is among the popular research directions in the field of automotive industry in recent years, and its emergence sets a trend for a new wave of automotive technology. Intelligent vehicles have the ability of autonomous perception, decision-making and control, and are able to realize a safe, efficient and comfortable driving experience by sensing the environment and making decisions according 1 . Lateral motion control for autonomous driving is one of the three main cores of this technology. According to the target path information from the upper-level decision planning system, corresponding steering control commands are output to control the vehicle along the target path. As being the core of the overall movement control system, the strengths and weaknesses of the transverse motion control methods will not only affect the tracking accuracy of the intelligent vehicle on the target path, but also have an impact on the stability and comfort of the whole vehicle 2 .
To date, many scholars have combined the design of intelligent driving vehicle systems with vehicle dynamics models to carry out research on lateral motion control methods, and have achieved fruitful results. Pereira 3 and others proposed a new nonlinear curvature response model of vehicles based on the MPC (Model Predictive Control) lgorithm, which was estimated online by Kalman filtering, and the designed controllers and curvature response models have high lateral control accuracy and stability, but the method requires high accuracy of the model and the precision of the environment perception, and it also requires real-time adjustment and optimization of the controller parameters, in addition, the computational complexity of the method is relatively high, and it requires a high-performance computing platform to achieve real-time performance and efficiency. Guo 4 designed a novel robust H ∞ fault-tolerant state feedback lateral control law, the controller remains capable of stabilizing the vehicle’s lateral motion and tracking the desired road despite actuator failures and parameter uncertainties. However, no real vehicle experiments were conducted to verify it. Hwang 5 proposed a robust nonlinear control method for lateral control of an autonomous vehicle using the barrier Lyapunov function (Barrier Lyapunov Function, Abbreviated as BLF) under the constraints of lateral offset error, which showed high robustness and stability in the face of lateral offset error and external disturbances, and was able to limit the lateral offset error so that the vehicle could accurately track the intended trajectory. Kang 6 proposed an improved ADRC(Active Disturbance Rejection Control) path tracking control method, which can estimate and compensate the vehicle dynamic characteristics and environmental changes in real time, and improve the robustness and stability of path tracking. Wang 7 designed an improved LQR (Linear Quadratic Regulator) controller based on the real-time updating algorithm and fuzzy control, and the experimental results show that the improved LQRcontroller is more robust and stable when facing the lateral offset error and external disturbances, so that the vehicle can accurately track the intended trajectory, and it is more robust and stable when facing the transverse offset error and external disturbances. The results show that this improved LQR lateral path tracking method has higher path tracking accuracy and stability, but still lacks in robustness. Literature on linear quadratic regulators 8 , 9 , 10 , 11 , 12 , 13 designed and tested lateral controllers for intelligent vehicles based on linear quadratic regulator (LQR), MPC, NMPC (Nonlinear Model Predictive Control), LPV (Linear Parameter-Varying) control method, LMI (Linear Matrix Inequality) robust control method, sliding film variable structure, PID (Proportional-Integral-Derivative Controller), and other control methods, respectively, and achieved good control effect scenarios in specific tests. In summary, the design of control methods is the core of the whole control system of the lateral control of intelligent vehicles. At present, the research on the control method mainly focuses on a single factor and has achieved certain results.
LQR control is one kind of optimal control, which has been applied in many fields 14 . In 1960, Rudolph E. Kalman proposed LQR for the first time, which laid the foundation of LQR theory 15 . Subsequently, the LQR theory was further improved by C Gokcek, PT Kabamba et al. It mainly includes the solution method of optimal state feedback control and the analysis of asymptotic stability 16 , 17 . In 1992, Hauser et al. proposed an optimal control method based on LQR applied to the vehicle formation control problem, and analysed its performance in maintaining the formation formation and reducing the following error, and achieved excellent control results 18 . Pacejka et al. proposed an active front wheel steering control strategy based on LQR in 1997, aiming to improve vehicle handling and stability, and the results showed that the LQR controller was excellent in handling nonlinear dynamics and uncertainty 18 , 19 . In 2009, Snider 20 of Stanford University proposed to apply the LQR control method to the path tracking control of a vehicle and modeled the dynamics with the curvature perturbation of the road surface included. Although this control method stabilizes the system, it does not take into account the effect of the disturbance term, and the tracker suffers from steady-state error. Kapania 21 proposed a feedforward + feedback LQR steering controller. The design first considered the nonlinear vehicle dynamics modeling and the construction of the controller for the feedforward control method. Meanwhile, it exhibits a significant increase in steady-state path deviation at high speeds. Xu et al. 22 proposed to add a road curvature feed-forward link on top of the LQR feedback control to eliminate the steady-state error of lateral displacement. In summary, the advantage of LQR in intelligent driving lateral control is that it can ensure that the intelligent vehicle tracking control system is close to the equilibrium state while consuming the minimum energy when the vehicle is yawing during driving. Research based on the LQR control method has been widely applied to intelligent vehicles, but control stability still needs to be improved.
Addressing the accuracy and stabilization problems of intelligent driving lateral motion control, and considering the interference factors of the practical environment, the paper proposes a feedforward + predictive LQR algorithm lateral motion control method based on Genetic Algorithm (GA) parameter optimization and PID steering angle compensation (referred to as FP + PID-GA-LQR in later Figs. 9 and 10). Firstly, a predictive controller is designed to be added to the traditional LQR controller to eliminate the system response lag problem. Secondly, a genetic algorithm is introduced for parameter optimisation, so that the control system is capable of adaptively updating the control parameters according to different driving environments. Then, a PID steering angle compensation controller is designed to compensate for the model mismatch due to the small angle assumption during the vehicle modelling process, which enhances the robustness of the system. Finally, the control algorithm is simulated in double lane change condition and circular condition by Carsim-Sinmulink joint simulation platform, and the simulation results are compared and analysed to verify the effectiveness of the control method proposed in this paper.
Establishment of vehicle dynamics model
Vehicle dynamics tracking error model.
Since the vehicle system is a complex nonlinear and time-varying parameter uncertainty system 23 , in the lateral control of the vehicle, the research mainly focuses on the lateral dynamics of the vehicle. In this paper, only two degrees of freedom of motion, lateral and transverse, are considered, and the vehicle is simplified as a two-degree-of-freedom “bicycle” model 24 , and the longitudinal velocity v x is assumed to be a constant as shown in Fig. 1 . Some of the structural parameters of the selected model based on Carsim are shown in Table 1 .
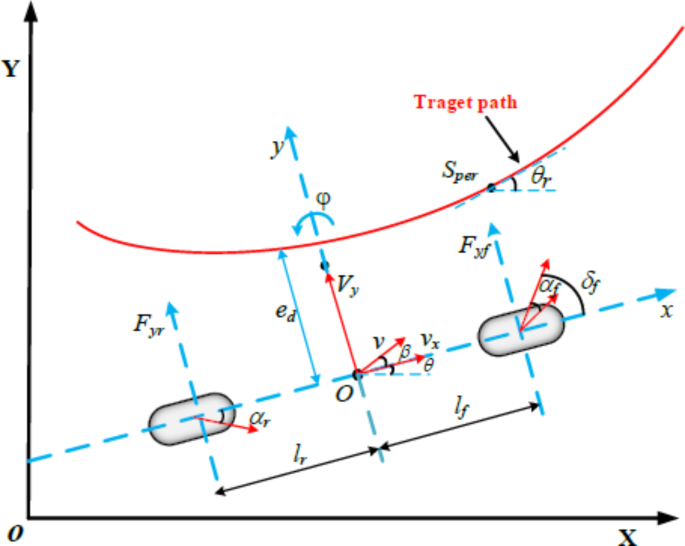
Vehicle dynamics tracking error model.
The model is based on the assumptions set out below:
The vehicle is traveling on a flat road; i.e., the vertical motion of the vehicle is not considered;
Without considering the influence of the vehicle suspension system;
The steering angle works directly on the front wheels of the vehicle;
The tires are in good contact with the ground;
Load transfer is neglected;
Air resistance is neglected.
By analyzing the two-degree-of-freedom vehicle model of Fig. 1 , assuming a constant vehicle speed and making small angle assumptions, the following dynamic equations can be obtained:
where m is the mass of the vehicle; F yf and F yr are the lateral forces on the front and rear axles of the vehicle, respectively; I z is the rotational moment of inertia of the vehicle around the Z-axis; \(\ddot {\varphi }\) is the angular acceleration of the vehicle’s transverse swing; l f and l r are the distances from the center of mass of the vehicle to the front and rear axles, respectively.
High-speed moving vehicles generate a lateral deflection angle in their tires. Experimental results indicate that at small deflection angles, the lateral force of the tire is directly proportional to the deflection angle, i.e. F y = Cα . Drawing on rigid body kinematics, the expressions for the lateral deflection angles of the front and rear wheels can be obtained as:
where β is the center of mass lateral deviation angle; δ f is the front wheel turning angle of the vehicle; the front wheel turning angle is generally small, so this paper makes tan( δ f + α f ) = δ f + α f , v x and v y represent the longitudinal velocity and lateral velocity of the car, respectively; \(\dot {\varphi }\) is the vehicle’s transverse pendulum angular velocity. Due to the existence of the transverse swing angle ϕ , the lateral acceleration of the vehicle \({a_y}={\dot {v}_y}+{v_x}\dot {\varphi },\) which is brought into the above kinetic equations, can be obtained by organizing the kinetic equations to obtain the state space expression:
Vehicle generates lateral and heading errors when tracking the planned path in real time. In practical control, in order to realize accurate lateral control, the designed lateral controller is required to eliminate these two errors in real time. Therefore, it is necessary to convert the vehicle’s dynamics model into a dynamics error model. The lateral error e d is defined as the distance between the center of mass of the vehicle and its projection point on the planning path; the heading error e ϕ is the deviation of the heading angle of the current position from the tangent direction of the projection point on the planning path.
According to the definition of lateral error e d , it can be expressed as:
where v is the center of mass velocity of the vehicle; θ is the heading angle of the vehicle; θ r is the theoretical heading angle at the current moment.
According to the definition heading angle is the sum of the transverse pendulum angle ϕ and the center of mass lateral deflection angle β . From the dynamics model in Fig. 1 , we can get v x = vcos ( β ), v y = vsin ( β ). Defining e ϕ as the approximate heading error and assuming that the difference between ϕ and θ r is particularly small, i.e., the value of β is small, then e ϕ = ϕ − θ r . After calculations and simplified and brought into Eqs. ( 2 ) and ( 3 ) can be obtained:
According to the definition of heading error e ϕ with the above analysis, it can be obtained:
Derivation of ( 7 ) can be launched:
Bringing Eq. ( 4 ) into Eqs. ( 6 ) and ( 8 ) yields \({\ddot {e}_d}\) :
Continued decomposition yields \({\ddot {e}_\varphi }\) :
Continuing the transformation of the above equations, the state space expressions for the lateral error and heading error of the vehicle while performing path tracking are obtained:
A rewriting of the above Eq. ( 11 ) yields a space state expression for the dynamics error model of the vehicle as:
As in real applications, especially in computers, discrete data are handled and the planned path is often tracked based on planned target points rather than a continuous path. So it is necessary to use discrete LQR control. Discretization of the above continuous state space equation ( 12 ), ignoring the effect of \({\dot {\theta }_r}\) , and integrating both sides of the equation yields:
By the median theorem of integration:
Generally speaking the accuracy of the system employing the midpoint Euler method is higher, but as the inputs are only known at the current moment, a mixture of the forward Euler method and midpoint Euler method is used for the above Eq. ( 14 ), so that \(X(\xi )=\left( {X(t)+X(t+dt)} \right)/2,U(\xi )=U(t)\) , substituting into Eq. ( 14 ), and the state-space equations of the discretized vehicle tracking error model can be obtained by organizing and simplifying as follows:
where \(\bar {A}={\left( {I - \frac{{Adt}}{2}} \right)^{ - 1}}\left( {I+\frac{{Adt}}{2}} \right),\bar {B}=Bdt\) and I denotes the unit matrix.
Lateral control system design
According to the error tracking model of the intelligent driving vehicle, the lateral controller illustrated in Fig. 2 as follows is designed to limit the lateral error and heading error of the vehicle during the target path tracking process, so as to minimise the state error between the vehicle’s current driving position and the reference position as much as possible.
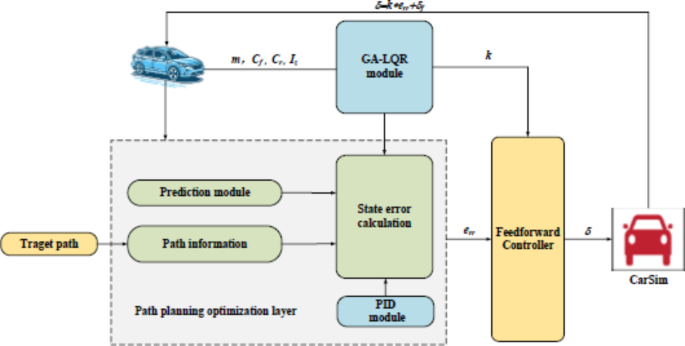
Design of lateral control system.
Initially, the vehicle parameters are output using the vehicle dynamics tracking error model established as above, the LQR controller is designed and discretised, and subsequently, the feedforward controller is devised to eliminate the steady state error of the system. Moreover, based on this, a predictive controller was added to eliminate the system lag. And the efficient and parallel global search capability of genetic algorithm (GA) is utilised to perform parameter optimisation of the weights of Q and R of the LQR controller based on the current target path. Then the target path of the vehicle to be simulated is selected and designed and combined with the feedforward controller to output the current position information of the vehicle and calculate the state error. Considering the ideal neglect assumptions in establishing the vehicle tracking model, and the design of the controller also relies on the simplified model, this paper uses a non-model-based PID controller to directly control and correct the transverse error, which compensates the final output of the front wheel angle by a small angle Δ δ . The feedforward controller is then combined with the GA controller, which is a feedforward controller, to calculate the state error of the vehicle, and the final output of the vehicle position information. Thereafter, the feedforward controller combined with the GA-LQR control module calculates and inputs the current desired front wheel steering angle of the vehicle. Ultimately, the output front wheel steering angle from the calculation module is fed back to the vehicle controller for real-time control to enable the vehicle to accurately track the target path.
Design of LQR controller
In modern control theory, a linear system represented by a state space expression is the mainly research object of LQR (Linear Quadratic Regulator), and the objective function is a quadratic function of the object function and control 25 . The advantage of Linear Quadratic Optimal Control (LQR) in the field of intelligent driving is that when the system has control deviations, it can use a small amount of control energy to make the system quickly approach the target state, which is not only stable, but also the control effect is fast. It is suitable for addressing multivariate, linear and linearisable full-state feedback control problems, and has good robustness against the effects of noise. Therefore, based on the discretised vehicle dynamics state space expression constructed by Eq. ( 15 ) in the previous section, the performance index function of the LQR controller is constructed as follows:
where X k is the state variable of the system; U k is the control variable of the system; Q and R represent the state error weighting matrix and the weighting matrix of the control quantity of the controller, respectively, where Q is a positive definite or semi-positive definite matrix and R is a positive definite matrix.
The essential core of the LQR controller is to design the control law such that Eq. ( 16 ) is minimised. Thus, the Hamiltonian function is constructed:
This is obtained by taking the derivative of Eq. ( 17 ) and resolving the extremes:
where λ k+1 = 2 P k+1 X k+1 is the solution of the Riccati equation P(t)PA + A T P-PBR −1 BP T + Q = 0.
As a corollary, the final control law of the LQR controller for a smart driving car is as follows:
where k = [k1, k2, k3, k4] is the gain of the LQR controller.
Feedforward controller design
By combining Eq. ( 19 ) equivalent Eq. ( 12 ) can be obtained:
By analysing Eq. ( 20 ), it can be observed that the system has a steady state error, i.e., it is not guaranteed that the lateral error and heading error of the vehicle during driving is 0. Therefore, in this paper, the design of a feedforward controller is considered to eliminate the steady state error. Setting the front wheel turning angle of the feedforward control output as δ k , the complete control law can be expressed as:
When the controller reaches the optimal control, i.e., when the steady state error of the system is 0, Eq. ( 21 ) can be obtained by bringing Eq. ( 12 ) into Eq. ( 12 ):
Laplace transforming Eq. ( 22 ) and solving for the extremes yields the steady state errors for lateral deviation e d_s and heading deviation e ϕ_s , respectively:
Through Eq. ( 23 ), it can be seen that the steady state error ed_s of the lateral deviation can be compensated and thus eliminated by feedforward control, while considering the assumptions made above e θ = ϕ + β − θ r = 0, and combining with Eq. ( 23 ) the following equation can be deduced:
According to Eq. ( 24 ), e θ = ϕ + β − θ r = 0, so the steady state error value of the heading error will not need the feedforward part for cancellation. In summary, the feedforward control law obtained in this question is:
where ρ denotes road curvature.
Predictive controller design
Humans are capable of making predictive driving manoeuvres based on changes in the environment and path conditions in front of them while driving a vehicle, however, the aforementioned LQR controller and feedforward controller that are designed based on the vehicle dynamics tracking error model do not possess the predictive performance. In order to enable vehicles to possess the capability of predicting future target paths and making driving actions in advance, Takemura et al. 26 designed long horizon predictive planning-tracking controllers by introducing position-field-of-view prediction similarity judgement method in the multi-intelligence path planning-tracking problem, which improved the planning and tracking performance of the intelligences in variable dynamic environments.
According to the test, it is observed that when the vehicle is in the situation shown in Fig. 3 , the lateral control system detects that there is a distance error between the vehicle and the target path, and the system determines that the steady state error is not zero in determining that the vehicle can not track the point S pre , but in the period of time after the point S pre of the target path, the vehicle is still capable of accurately tracking the target path without changing the angle of rotation of the front wheels.
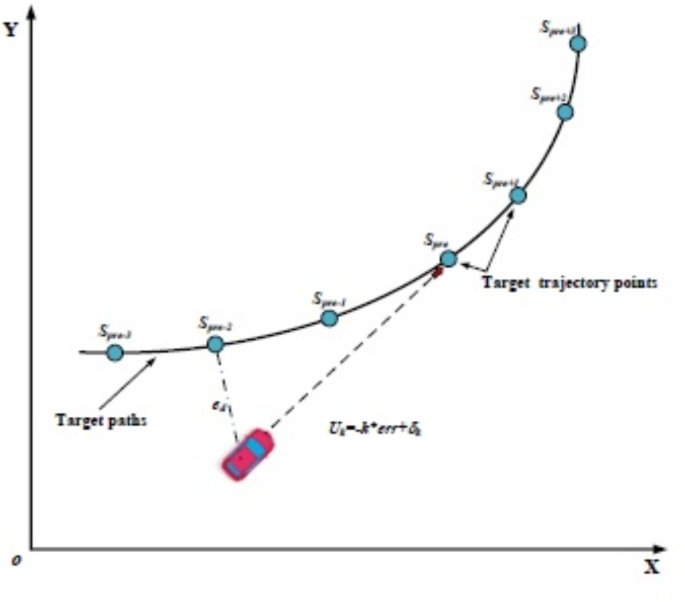
Schematic diagram of scenario I.
As shown in Fig. 4 , the vehicle is currently travelling on the target path, at which the lateral control system detects that the lateral error is zero and the heading error is close to zero, i.e., the system determines that the steady-state error is zero and makes the determination that the vehicle is able to track the target path point. Until the vehicle gradually deviates from the target path, the system will not control the vehicle until it detects the steady state error, so that the vehicle can keep travelling on the target path point S pre_t2 , which is caused by the control hysteresis of the feedforward-LQR controller.
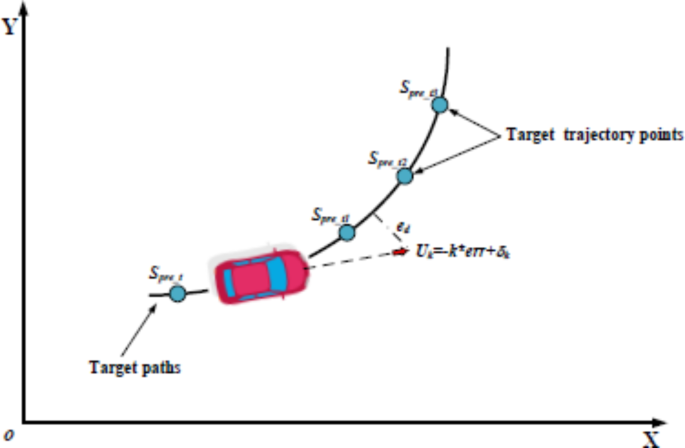
Schematic diagram of scenario II.
Aiming at the above possible issues, this paper adds a predictive controller on the basis of the feedforward-LQR controller, Fig. 5 illustrates the predictive control model, from which it can be seen that it eliminates the lag of the control system by predicting the target path points in the future period of time, and then the lateral control system makes motion control operations on the vehicle in advance. This improves the operating efficiency of the control system, which in turn improves the path tracking accuracy and driving stability as well as comfort.
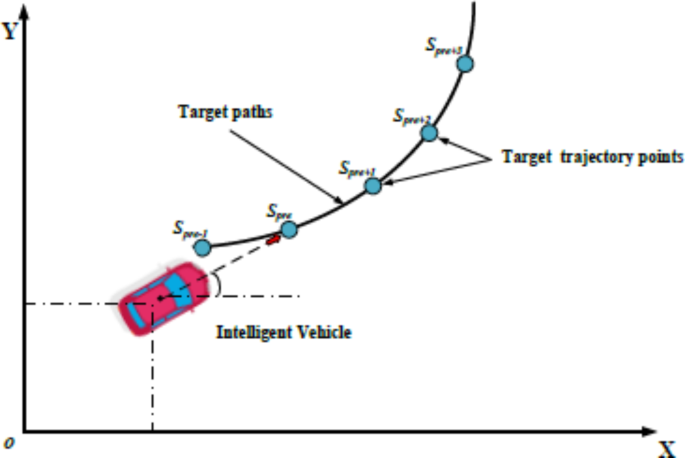
Schematic diagram of the predictive control model.
The prediction time is setting as ts and the coordinate location information of the prediction time points are ( X pre , y pre , v xpre , v ypre , φ pre , \({\dot {\varphi }_{per}}\) ) Based on the θ = ϕ + β menioned above, the following equation can be obtained.
Design of PID steering angle compensation controller
Several simplifying assumptions are applied in constructing the vehicle error tracking model. Although feedforward control can reduce the steady state error to some extent, it is considered that in practice there are errors in terms of model mismatch and tyre side bias stiffness estimation. In addition, the controller design also relies on simplified models and feedforward control may face a number of problems such as hysteresis control. Therefore, in this paper, a PID controller that is not model-based is employed to directly control the correction of lateral errors. In this way, small-angle Δδ steering compensation is performed and the corrected result is compensated into the front wheel steering angle to improve the effect of steady state error caused by model mismatch and inaccurate parameter estimation. Figure 6 below shows the technology roadmap for PID steering compensation controllers, with the transfer function between G(s) inputs and outputs.
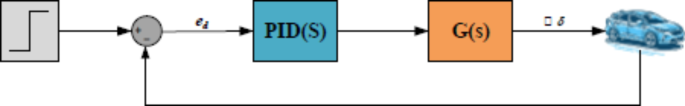
Technical route of PID steering compensation controller.
where Δ δ is the small angle steering angle compensation; a 1 , a 2 , a 3 are the proportionality coefficient, integration coefficient, differential coefficient, respectively.
The performance of the PID controller depends on its three tuning parameters namely proportional, integral and differential gain. In this paper, parameter tuning has been carried out with reference to a univariate tuning parameter formula for online robustness and performance tuning proposed by Verma et al. 27 . The obtained parameter results are shown in Table 2 below.
Genetic algorithm for LQR controller parameter optimisation
As the designed LQR transversal controller is applied in path tracking, the weight matrices Q and R in the LQR control algorithm take values that have a significant impact on the control performance of this algorithm. In case Q is larger, the control algorithm performs better but at the expense of stability. In case R is larger, the control process of tracking will be smoother and smoother, and the angle of the front wheel will not change drastically, which ensures the safety of the system, but it is easily caused by the problem of reduced tracking accuracy. The state weight matrix Q and control energy weight matrix R of the traditional LQR controller are often based on experience, relying on manual step-by-step trial and error, which is highly subjective. Therefore, in this paper, we consider introducing a genetic algorithm with better global search capability to solve the optimisation problem.
The general structure of a genetic algorithm is presented in Fig. 7 . To begin with, a set of initial populations is randomly generated, and then new populations are continuously generated by selecting, crossing over and mutating 28 . Afterwards, the parameters are continuously modified by minimising the fitness factor until a fixed maximum generation is obtained.
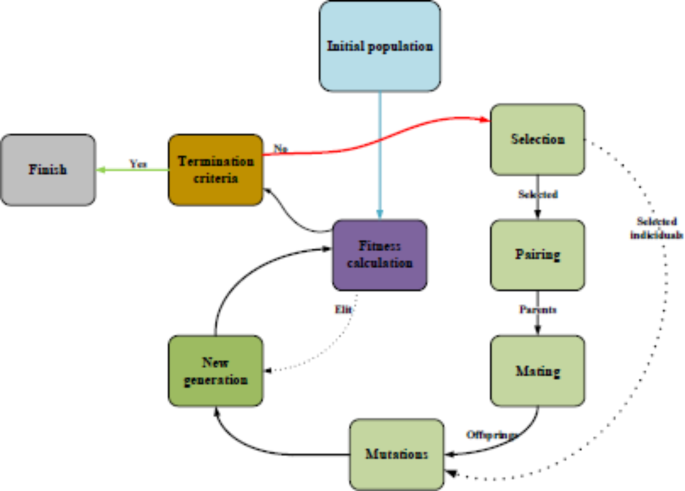
Generic architecture of GA.
The specific parameters of the genetic algorithm (GA) are shown in Table 3 , where both Q and R are diagonal matrices, and the appropriate variables are selected for focused optimisation based on the importance of the relevant variables to the research objectives, as a way to improve the performance and computing speed of the controller. At the same time, in order to make the initial solution uniformly distributed in the search space, this paper sets an appropriate population size.
The evolutionary direction in genetic algorithm is determined by the calculation of the adaptation degree. Considering the control accuracy and vehicle stability, this paper designs the performance index function P GA and the fitness function F , which combines the effects of Q and R . The lateral error ed, the heading error e ϕ and the front wheel angle δ f in the path tracking are also taken into account, furthermore, these variables are normalised by a fixed mean obtained from a single test in order to reduce the size differences.
where η 1 , η 2 , and η 3 are weighting factors characterising the importance given to lateral error, heading error, and front wheel angle respectively, and the sum of the three is one.
The selection operation in genetic algorithms essentially embodies a process of natural selection, i.e., individuals that are more adaptable to the environment have a greater probability of being inherited by the following generation, while individuals with weaker adaptive ability have a lower probability of being inherited by the next generation. In order to select excellent individuals as parents to generate new populations, and to better adapt to the problem characteristics and optimisation objectives of path tracking, this paper chooses to employ roulette selection and uniform sampling 29 , which avoids certain individuals from monopolising the selection process, and thus improves the fairness of the original roulette selection.
According to the above parameters and the established fitness function, the execution is looped until the termination criterion set in this paper is reached, and the current global optimal solution and LQR optimal weight matrix are output. Figure 8 illustrates the relationship between the fitness value and the number of iterations for the following two simulation conditions, with Q and R weights as shown in Table 4 below.
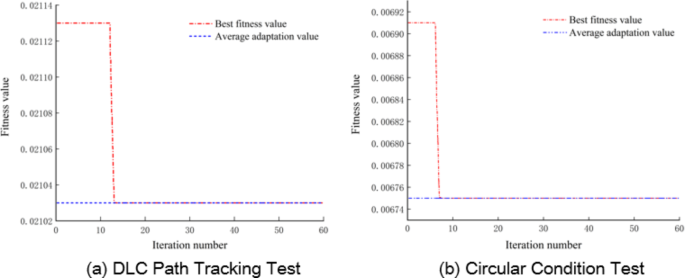
Relationship between fitness value and number of iterations.
Simulation analysis
Dlc path tracking test.
Simulation validation is carried out based on the Carsim-Simulink co-simulation platform, and the simulation conditions are selected as double lane change conditions (DLC), the road adhesion coefficient µ is divided into two sections: 0.5 and 0.8, with road surface roughness (IRI) of 2 m/km and 5 m/km, respectively. In order to better evaluate the performance of the feedforward + predictive LQR algorithm controller based on PID steering angle compensation and GA parameter optimisation designed in this paper, the simulation results are tested in comparison with those of the feedforward-control-free LQR controller and the feedforward + LQR controller were compared and tested with the two LQR controller weight coefficients Q = [30,1,5,1], R = 6. The test results are shown in Fig. 9 below.
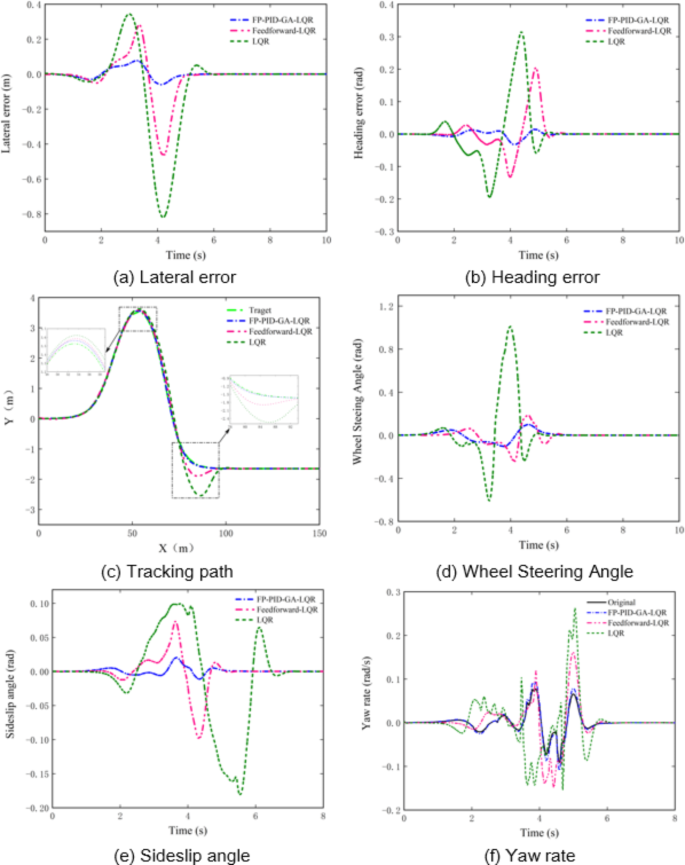
Double lane change condition test results (72 km/h).
Figure 9 c demonstrates the comparison of the tracking effect of the three controllers under the double lane change line condition at a speed of 72 km/h. As can be seen from the figure, all three controllers are able to track the target path effectively from the perspective of the whole path, and the feedforward-predictive + PID-GA-LQR control has the most efficient tracking effect on the overall path of the vehicle, particularly in the two c feedforward-predictive + PID-GA-LQR urves of the target path, which is more efficient compared to the other two controllers. Figure 8 a and b show the lateral error and heading error of the three controllers under this working condition. It can be observed that in the straight line phase at the beginning and near the end of the target path, the three control methods can track the target path better, and the lateral error and heading error can be controlled within a more reasonable range, but the LQR controller without added feedforward control has an obvious hysteresis phenomenon in the control of the vehicle compared to the feedforward + LQR controller and the feedforward-predictive + PID-GA-LQR controller, Which also caused a remarkable variation in the adjustment of the amount of error by the three controllers. From Table 5 , it can be observed that the maximum lateral error and the maximum heading error of the feedforward-predictive + PID-GA-LQR controller are 0.077 m and 0.033 rad, respectively, the control effect of the feedforward + LQR controller is relatively poor, with the maximum lateral error value reaching 0.465 m, and the maximum value of the heading error reaching 0.204 rad, and the control effect of the LQR controller is the worst among all three controllers, and the maximum lateral error has reached 0.204 rad. The maximum lateral error has reached 0.820 m, and the maximum value of heading error has reached 0.314 rad. Figure 9 d shows the variation of the front wheel angle during tracking of the three controllers, from the figure, it can be observed that the feedforward-predictive + PID-GA-LQR controller outputs a smoother front wheel angle during the control process, and the overall fluctuation is the smoothest compared to the feedforward + LQR controller and the LQR controller, and Fig. 4 b also shows that in the simulation condition from 2 to 3 s, the heading errors of the feedforward + LQR controller and the LQR controller change frequently, and the fluctuation is more obvious and drastic.
Generally speaking, under the 72 km/h double lane change test condition, the feedforward-predictive + PID-GA-LQR controller improves 83.4% and 83.8% in terms of the maximum error control accuracy of the lateral error and heading error compared to the feedforward + LQR controller, and 90.6% and 89.4% compared to the LQR controller. And from Fig. 9 e and f, it can be known that the vehicle sideslip angle of the proposed control method in this paper is significantly reduced compared with the feedforward-LQR and LQR controllers, which is only between − 0.05° and 0.05°, and the vehicle yaw rate ranged from − 0.15 rad/s and 0.1 rad/s, which is obviously closer to the original value compared with the other two control strategies. The results indicate that the controller has more excellent tracking performance during the whole tracking process, and the stability of the vehicle is significantly improved.
Circular condition test
Circular conditions are a common scenario in automobile driving, which involves steering, acceleration and deceleration and various other driving skills, and changing the environmental factors (e.g., wind speed, friction coefficient, etc.) in the circular conditions test could be a great way to test the robustness and reliability of the path-tracking controller. Therefore, in this paper, the circular condition is used in addition to the double lane change condition for simulation testing. Considering the actual driving situation, the test speed is set to 40 km/h. The road adhesion coefficient µ is divided into two sections: 0.4 and 0.7, with road surface roughness (IRI) of 2 m/km and 6 m/km, respectively. The test results are shown in Fig. 10 below.
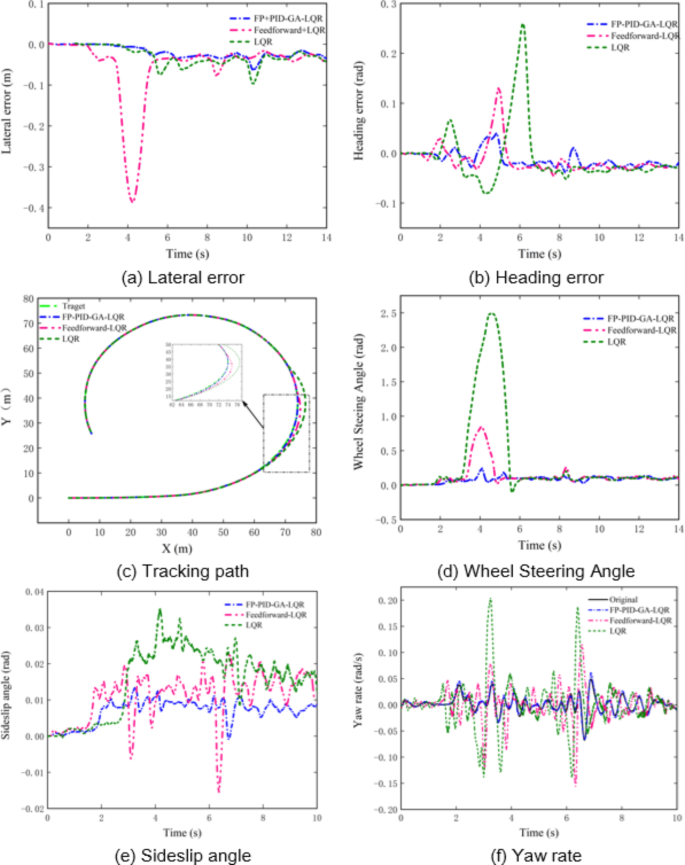
Circular condition test results (40 km/h).
Figure 10 c demonstrates a comparative graph of the tracking performance of the three controllers for a circular condition with a speed of 40 km/h. As can be seen from the figure, all three control methods are able to track the target path better in the starting phase, yet after entering the curved path, all three controllers appear to gradually deviate from the target path, with the feedforward-predictive + PID-GA-LQR control having the relatively smallest amount of vehicle deviation. Figure 8 a and b demonstrate the lateral error and heading error of the three controllers under this operating condition. It can be observed that in the straight line stage at the beginning of the target path, each of the three control methods can track the target path well, but upon entering the curved path, the feedforward + LQR controller and the LQR controller, which have not been optimised for the GA parameters and compensated for the PID steering angle, show large lateral and heading errors, and the LQR controller, which has not been added with the feedforward, shows hysteresis, but due to the low speed, the hysteresis degree is improved relative to the 72 km/h double lane change condition. From Table 6 , it was observed that the maximum lateral error and the maximum heading error of the feedforward-predictive + PID-GA-LQR controller were 0.053 m and 0.043 rad, respectively, and the control effect of the feedforward + LQR controller was relatively poor, with the maximum value of the lateral error exceeding 0.108 m, and the maximum value of the heading error exceeding 0.119 rad, and the LQR controller remained the worst control effect among all three. effect is the worst, with the maximum lateral error value exceeding 0.386 m and the maximum value of heading error exceeding 0.260 rad. Figure 10 d demonstrates the variation of the front wheel angle during tracking for the three controllers. It can be seen from the figure that the overall fluctuation of the front wheel steering angle output from the feedforward-predictive + PID-GA-LQR controller is smoother compared to the feedforward + LQR controller and the LQR controller during the tracking process of the curved path.
In addition, it is observed in Table 6 that the feedforward-predictive + PID-GA-LQR controller improves 51.9% and 63.8% in terms of the maximum error control accuracy of lateral error and heading error compared to the feedforward + LQR controller, and 85.4% and 85.0% compared to the LQR controller, respectively. As can be seen in Fig. 10 e and f, the vehicle sidelip angle and vehicle yaw rate of the controllers designed in this paper range from − 0.01° to 0.02° and from − 0.10 rad/s to 0.05 rad/s, respectively, which are significantly reduced compared to both the feedforward-LQR and LQR controllers. The results indicate that the feedforward- predictive LQR controller with GA parameter optimisation and PID steering angle compensation achieved the most effective tracking effect and higher stability of the vehicle driving process under the 40 km/h circular path test conditions.
Conclusions
The lateral motion control problem of intelligent vehicles has been investigated for one of the core in the intelligent vehicle autopilot structure system. Above all, based on the establishment of vehicle dynamics tracking error model, according to the linear quadratic regulator (LQR) control principle, the LQR lateral motion controller for intelligent vehicles was designed, and at the meantime, the feedforward controller was designed for the existence of steady state error in the system in an aim to eliminate the steady state error. Then, a predictive controller was added to the feedforward LQR controller to eliminate system lag.Subsequently, PID steering angle compensation controller was designed to take advantage of its non-model-based advantage to directly control and correct the transverse error, compensating for the effects of steady state errors that may occur caused by model mismatch and inaccurate parameter estimation. Secondly, to address the problems of cumbersome parameter adjustment of the LQR controller and easy mismatch of the fixed parameters with the model, A path-tracking error based LQR controller is devised by exploiting the global search capability of Genetic Algorithm (GA), which optimises the control parameters of the transverse motion controller and improves the adaptivity of the control accuracy. In the end, the simulation verification and comparative analysis were conducted under two working conditions using the Carsim- Simulink joint simulation platform. The simulation results indicate that, compared with the primitive LQR controller with fixed weight values, the lateral motion controller for intelligent vehicles designed in this paper has more good path tracking accuracy and vehicle stability, as well as more excellent convergence speed, dynamic performance and response time. Considering that the controller proposed in this paper does not take into account the effect of longitudinal velocity changes, the following step will be planned to design a combination of lateral_ longitudinal coupling control to establish a more improved path-tracking control system for ntelligent vehicle.
Data availability
The data that support the findings of this study are available from the corresponding author, [Zimo Ye], upon reasonable request.
Giaimo, F. & Berger, C. Design criteria to architect continuous experimentation for self-driving vehicles. Arxiv preprint. arXiv:1705.05170 (2017).
Xin, M. & Minor, M. A. Variable structure backstepping control via a hierarchical manifold set for graceful ground vehicle path following. In Robotics and Automation (ICRA), 2013 IEEE International Conference on 2013.
Pereira, G. C., Wahlberg, B., Pettersson, H. et al. Adaptive reference aware MPC for lateral control of autonomous vehicles. Control Eng. Pract. https://doi.org/10.1016/j.conengprac.2022.105403 (2023).
Article Google Scholar
Guo, J., Luo, Y. & Li, K. Robust H ∞ fault-tolerant lateral control of four-wheel-steering autonomous vehicles. Int. J. Automot. Technol. 21 , 993–1000. https://doi.org/10.1007/s12239-020-0094-8 (2020).
Hwang, Y., Kang, C. M. & Kim, W. Robust nonlinear control using barrier lyapunov function under lateral offset error constraint for lateral control of autonomous vehicles. IEEE Trans. Intell. Transp. Syst. 23 , 1565–1571. https://doi.org/10.1109/tits.2020.3023617 (2022).
Kang, N. et al. Improved ADRC-based autonomous vehicle path-tracking control study considering lateral stability. Appl. Sci. Basel. https://doi.org/10.3390/app12094660 (2022).
Wang, Z. et al. Improved linear quadratic regulator lateral path tracking approach based on a real-time updated algorithm with fuzzy control and cosine similarity for autonomous vehicles. Electronics . https://doi.org/10.3390/electronics11223703 (2022).
Article PubMed PubMed Central Google Scholar
Cui, Q. et al. Path-tracking and lateral stabilisation for autonomous vehicles by using the steering angle envelope. Veh. Syst. Dyn. 59 , 1672–1696. https://doi.org/10.1080/00423114.2020.1776344 (2021).
Article ADS Google Scholar
Li, P. et al. Polytopic LPV approaches for intelligent automotive systems: State of the art and future challenges. Mech. Syst. Signal Process. https://doi.org/10.1016/j.ymssp.2021.107931 (2021).
Guo, N. et al. A real-time nonlinear model predictive controller for yaw motion optimization of distributed drive electric vehicles. IEEE Trans. Veh. Technol. 69 , 4935–4946. https://doi.org/10.1109/tvt.2020.2980169 (2020).
Tang, F. & Li, C. Intelligent vehicle lateral tracking control based on multiple model prediction. AIP Adv. https://doi.org/10.1063/1.5141506 (2020).
Du, G. et al. Hierarchical motion planning and tracking for autonomous vehicles using global heuristic based potential field and reinforcement learning based predictive control. IEEE Trans. Intell. Transp. Syst. 24 , 8304–8323. https://doi.org/10.1109/tits.2023.3266195 (2023).
Li, Y. et al. Intelligent PID guidance control for AUV path tracking. J. Cent. South. Univ. 22 , 3440–3449. https://doi.org/10.1007/s11771-015-2884-0 (2015).
Shauqee, M. N., Rajendran, P. & Suhadis, N. M. An effective proportional-double derivative-linear quadratic regulator controller for quadcopter attitude and altitude control. Automatika 62 , 415–433. https://doi.org/10.1080/00051144.2021.1981527 (2021).
Kalman, R. E. Contributions to the theory of optimal control (1960).
Gokcek, C., Kabamba, P. T. & Meerkov, S. M. SLQR/SLQG: An LQR/LQG theory for systems with saturating actuators. IEEE (2000).
Gokcek, Cevat, Kabamba, et al. An LQR/LQG theory for systems with saturating actuators. IEEE Transactions on Automatic Control 2001.
Amer, N. H. & Zamzuri, H. Modelling and Control strategies in path tracking control for autonomous ground vehicles: A review of state of the art and challenges. J. Intell. Robot. Syst. 86 , 1–30 (2017).
Falcone, P. et al. Predictive active steering control for autonomous vehicle systems. IEEE Trans. Control Syst. Technol. 15 , 566–580 (2007).
Snider, J. M. Automatic Steering Methods for Autonomous Automobile Path Tracking . Robotics Institute.
Kapania, N. R. & Gerdes, J. C. Design of a feedback-feedforward steering controller for accurate path tracking and stability at the limits of handling. Veh. Syst. Dyn. 53 , 1687–1704. https://doi.org/10.1080/00423114.2015.1055279 (2015).
Xu, S., Peng, H. & Design,. Design, analysis, and experiments of preview path tracking control for autonomous vehicles. IEEE Trans. Intell. Transp. Syst. 21 , 48–58. https://doi.org/10.1109/tits.2019.2892926 (2020).
Li, Y. Dynamic system optimal performances of shared autonomous and human vehicle system for heterogeneous travellers. Math. Comput. Model. Dyn. Syst. 26 , 481–499. https://doi.org/10.1080/13873954.2020.1792509 (2020).
Article MathSciNet Google Scholar
Cheng, S. et al. Longitudinal collision avoidance and lateral stability adaptive control system based on MPC of autonomous vehicles. IEEE Trans. Intell. Transp. Syst. 21 , 2376–2385. https://doi.org/10.1109/tits.2019.2918176 (2020).
Katebi, J. et al. Developed comparative analysis of metaheuristic optimization algorithms for optimal active control of structures. Eng. Comput. 36 , 1539–1558. https://doi.org/10.1007/s00366-019-00780-7 (2020).
Takemura, N. et al. A path-planning method for human-tracking agents based on long-term prediction. IEEE Trans. Syst. Man. Cybern Part. C Appl Rev. 42 , 1543–1554. https://doi.org/10.1109/tsmcc.2012.2203801 (2012).
Verma, B. & Padhy, P. K. Robust fine tuning of optimal PID controller with guaranteed robustness. IEEE Trans. Ind. Electron. 67 , 4911–4920. https://doi.org/10.1109/tie.2019.2924603 (2020).
Li, H. et al. Safety research on stabilization of autonomous vehicles based on improved-LQR control. AIP Adv. https://doi.org/10.1063/5.0078950 (2022).
Lipowski, A. & Lipowska, D. Roulette-wheel selection via stochastic acceptance. Phys. A Stat. Mech. Appl. 391 , 2193–2196. https://doi.org/10.1016/j.physa.2011.12.004 (2012).
Download references
Acknowledgements
All the authors would like to express our gratitude to all those who helped us improve this work.
National Natural Science Foundation of China, 51875494, Zhu’an Zheng; Natural Science Research of Jiangsu Higher Education Institutions of China, 19KJB580019, Zhu’an Zheng; Key University Science Research Project of Jiangsu Province, SJCX20_1353, Zhu’an Zheng; Yancheng institute of Technology Postgraduate Innovation Programme Project, SJCX23_XZ027, Zimo Ye.
Author information
Authors and affiliations.
School of Automotive Engineering, Yancheng Institute of Technology, No. 1 Middle Hope Avenue, Tinghu District, Yancheng City, 224000, Jiangsu Province, China
Zhu-an Zheng, Zimo Ye & Xiangyu Zheng
You can also search for this author in PubMed Google Scholar
Contributions
Author contribution: Conceptualization, Z.A.Z.; methodology, Z.A.Z.; software, Z.M.Y.; formal analysis, Z.M.Y.; Investigation, Z.M.Y.; resources, Z.A.Z.; data curation, X.Y.Z.; writing—original draft preparation, Z.M.Y.; writing—review and editing, Z.A.Z.; project administration, Z.M.Y.; funding acquisition, Z.A.Z., Z.M.Y.; Supervision, Z.A.Z.; Visualization, Z.M.Y. and X.Y.Z.; All authors have read and agreed to the published version of the manuscript.
Corresponding author
Correspondence to Zimo Ye .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
Zheng, Za., Ye, Z. & Zheng, X. Intelligent vehicle lateral control strategy research based on feedforward + predictive LQR algorithm with GA optimisation and PID compensation. Sci Rep 14 , 22317 (2024). https://doi.org/10.1038/s41598-024-72960-5
Download citation
Received : 29 May 2024
Accepted : 12 September 2024
Published : 27 September 2024
DOI : https://doi.org/10.1038/s41598-024-72960-5
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Autonomous driving
- Lateral motion control
- Genetic algorithm
- Linear-quadratic optimal control
- Path tracking
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

IMAGES
VIDEO
COMMENTS
Genetic testing may also be used as a method for the determination of parentage or ancestry. Latest Research and Reviews Nationwide survey of the secondary findings in cancer genomic profiling ...
In 1987, the New York Times Magazine characterized the Human Genome Project as the "biggest, costliest, most provocative biomedical research project in history." 2 But in the years between the ...
The widening scope and scale of genetic tests are posing new challenges, and the need to address these challenges is becoming immediately relevant for all clinicians, not just genetics experts. Notes
The 2024 Albert Lasker Award for Basic Medical Research recognizes Dr. Zhijian (James) Chen for his elucidation of how DNA stimulates immune responses. Correspondence Sep 18, 2024
Applied Clinical Genomics is the application of genetic information to the clinical setting, including improved diagnosis of disease and tailored treatment efficacy and safety. By discovering and defining the genes that underlie susceptibility to disorders, genetic information can be used to identify and better define those genes that play ...
Introduction. The past two decades have seen major shifts in our technical ability to sequence genetic information at scale. Historically, genetic testing tended to consist of either highly detailed molecular testing of nominated single genes, or broad genome-wide dosage screening at low resolution, for example karyotyping [1,2].Genome sequencing was too slow and too expensive to be used in ...
These include (1) a traditional genetic health-care model of services between genetics health-care providers and a patient's referring provider, (2) a nontraditional genetic health-care model ...
RSS Feed. Genetics research is the scientific discipline concerned with the study of the role of genes in traits such as the development of disease. It has a key role in identifying potential ...
Genetics is a branch of Biology to start with. It is mainly focused on the study of genetic variation, hereditary traits, and genes. Genetics has relations with several other subjects, including biotechnology, medicine, and agriculture. In Genetics, we study how genes act on the cell and how they're transmitted from a parent to the offspring.
Research methods: DNA sequencing and genomics: Genetic testing: Cell-free fetal DNA: Newborn screening: Diagnostic testing: Carrier testing: Preimplantation genetic diagnosis: Prenatal diagnosis: Predictive and presymptomatic testing: Pharmacogenomics: Non-diagnostic testing: Forensic testing: Paternity testing: Genealogical DNA test: Research ...
This page lists various research-topics about genetics to help researchers find relevant research articles. ... Genetic Testing In Treatment and Management of Alzheimer's Disease. Alzheimer's disease (AD) is a progressive neurodegenerative disorder that affects an estimated 50 million people worldwide. This disease is currently the most ...
Barriers presented by the current model. Barriers, as it applies to genetic screening and testing, refers to the individual, provider, system, and policy level hurdles an individual must overcome to access genetic testing and navigate the process of genetic testing. Investigators have done extensive work identifying the multilevel barriers ...
Decoding Complexity: Genomic, Epigenomic, and Environmental Dynamics in Developmental and Neurogenetic Disorders. Alessandro Fiorenzano. Luisa Azevedo. 338 views. The most cited genetics and heredity journal, which advances our understanding of genes from humans to plants and other model organisms. It highlights developments in the function and ...
Providers often cite lack of genetic knowledge/confidence as a reason for lack of testing or referring for genetic testing. 64, 65, 83 For non-geneticists, education efforts should include both "standard" and intensive training options, depending on providers' intentions to order/interpret/disclose testing on their own or alongside a ...
RSS Feed. Genetics is the branch of science concerned with genes, heredity, and variation in living organisms. It seeks to understand the process of trait inheritance from parents to offspring ...
213 Genetics Research Topics & Essay Questions for College and High School. Genetics studies how genes and traits pass from generation to generation. It has practical applications in many areas, such as genetic engineering, gene therapy, gene editing, and genetic testing. If you're looking for exciting genetics topics for presentation, you ...
Genetic/genomic testing (GGT) are useful tools for improving health and preventing diseases. Still, since GGT deals with sensitive personal information that could significantly impact a patient's life or that of their family, it becomes imperative to consider Ethical, Legal and Social Implications (ELSI). Thus, ELSI studies aim to identify and address concerns raised by genomic research that ...
In 2014, the Deciphering Developmental Disorders Study reported the first results from the sequencing of 12,000 children with developmental disorders 2. At the same time, work began on a project to sequence 100,000 genomes of 70,000 NHS patients 3. Outside of the UK, sequencing in healthcare settings is also expanding.
In celebration of the 20th anniversary of Nature Reviews Genetics, we asked 12 leading researchers to reflect on the key challenges and opportunities faced by the field of genetics and genomics.
Research is examining how germ cells are specified. We study the broad range of biology required to transmit genetic information from one generation to another, and how to facilitate the process of reproduction when difficulties arise or to avoid passing on mutant genes. Faculty use genetics and molecular genetic approaches to understand ...
122 The Best Genetics Research Topics For Projects. The study of genetics takes place across different levels of the education system in academic facilities all around the world. It is an academic discipline that seeks to explain the mechanism of heredity and genes in living organisms. First discovered back in the 1850s, the study of genetics ...
Genetic engineering is the act of modifying the genetic makeup of an organism. Modifications can be generated by methods such as gene targeting, nuclear transplantation, transfection of synthetic ...
A total of 3505 documents were published between 1952 and 2021, with a trend toward increase in paper/year. The most common documents are original articles (2515, 71.8%), followed by review articles (341, 9.7%). ... Genetic counseling researchers may use these keywords to find topics pertinent to their future research and practice. Subject ...
Explore 100+ detailed genetics presentation topics, from basic concepts to advanced research, genetic disorders, and ethical considerations. Perfect for any audience. Genetics is a fascinating field that explores the science of genes, heredity, and variation in living organisms.
This review paper covers the forensic-relevant literature in biological sciences from 2019 to 2022 as a part of the 20th INTERPOL International Forensic Science Managers Symposium. Topics reviewed include rapid DNA testing, using law enforcement DNA databases plus investigative genetic genealogy DNA databases along with privacy/ethical issues ...
Intelligent Vehicle is among the popular research directions in the field of automotive industry in recent years, and its emergence sets a trend for a new wave of automotive technology.